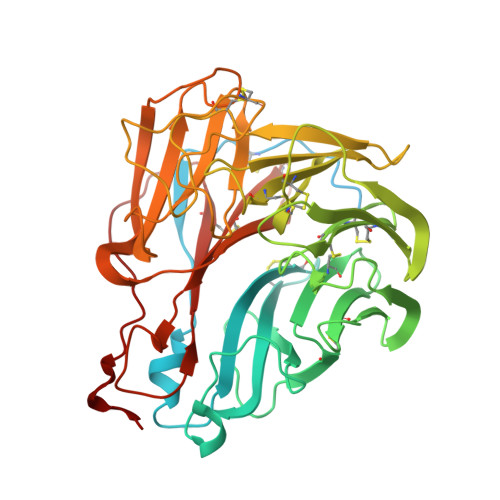
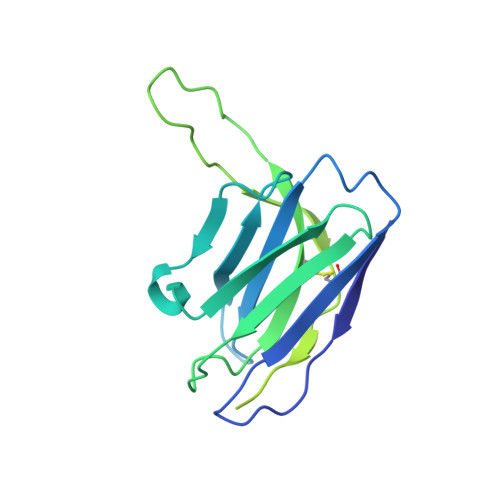
Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Lederhofer, J., Tsybovsky, Y., Nguyen, L., Raab, J.E., Creanga, A., Stephens, T., Gillespie, R.A., Syeda, H.Z., Fisher, B.E., Skertic, M., Yap, C., Schaub, A.J., Rawi, R., Kwong, P.D., Graham, B.S., McDermott, A.B., Andrews, S.F., King, N.P., Kanekiyo, M.(2024) Immunity 57: 574-586.e7
- PubMed: 38430907
- DOI: https://doi.org/10.1016/j.immuni.2024.02.003
- Primary Citation of Related Structures:
8GAT, 8GAU, 8GAV - PubMed Abstract:
Continuously evolving influenza viruses cause seasonal epidemics and pose global pandemic threats. Although viral neuraminidase (NA) is an effective drug and vaccine target, our understanding of the NA antigenic landscape still remains incomplete. Here, we describe NA-specific human antibodies that target the underside of the NA globular head domain, inhibit viral propagation of a wide range of human H3N2, swine-origin variant H3N2, and H2N2 viruses, and confer both pre- and post-exposure protection against lethal H3N2 infection in mice. Cryo-EM structures of two such antibodies in complex with NA reveal non-overlapping epitopes covering the underside of the NA head. These sites are highly conserved among N2 NAs yet inaccessible unless the NA head tilts or dissociates. Our findings help guide the development of effective countermeasures against ever-changing influenza viruses by identifying hidden conserved sites of vulnerability on the NA underside.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.