A hinge glycan regulates spike bending and impacts coronavirus infectivity
Chmielewski, D., Wilson, E., Pintilie, G., Chen, M., Schmid, M.F., Wells, L., Jin, J., Singharoy, A., Chiu, W.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
This entry reflects an alternative modeling of the original data in: 7kip
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike glycoprotein | A, B [auth C], C [auth B] | 1,356 | Human coronavirus NL63 | Mutation(s): 0 Gene Names: S, 2 | 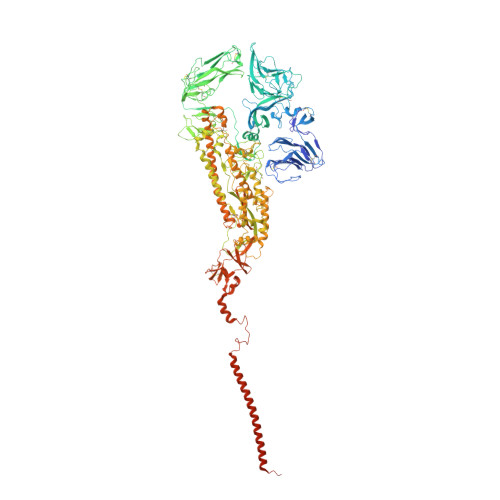 |
UniProt | |||||
Find proteins for Q6Q1S2 (Human coronavirus NL63) Explore Q6Q1S2 Go to UniProtKB: Q6Q1S2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6Q1S2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | AD [auth qA], BA [auth b], CB [auth 2], D, DB [auth 3], AD [auth qA], BA [auth b], CB [auth 2], D, DB [auth 3], EA [auth e], EB [auth 4], ED [auth uA], GD [auth wA], HB [auth 7], HC [auth XA], IA [auth i], ID [auth yA], KA [auth k], KB [auth AA], L, MA [auth m], MB [auth CA], NC [auth dA], OA [auth o], OC [auth eA], PB [auth FA], PC [auth fA], R, S, SC [auth iA], T, TB [auth JA], VB [auth LA], VC [auth lA], W, WA [auth w], XB [auth NA], XC [auth nA], Z, ZB [auth PA] | 7 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G97423PY GlyCosmos: G97423PY GlyGen: G97423PY | |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | AA [auth a], AB [auth 0], AC [auth QA], BB [auth 1], BD [auth rA], AA [auth a], AB [auth 0], AC [auth QA], BB [auth 1], BD [auth rA], CA [auth c], CD [auth sA], DC [auth TA], DD [auth tA], E, EC [auth UA], FA [auth f], FB [auth 5], FC [auth VA], GA [auth g], GB [auth 6], GC [auth WA], H, HA [auth h], HD [auth xA], I, IB [auth 8], J, JB [auth 9], JC [auth ZA], K, KC [auth aA], LA [auth l], LB [auth BA], LC [auth bA], MC [auth cA], N, NB [auth DA], O, P, PA [auth p], Q, QB [auth GA], QC [auth gA], RB [auth HA], RC [auth hA], SA [auth s], SB [auth IA], TA [auth t], TC [auth jA], U, UA [auth u], UC [auth kA], V, VA [auth v], WB [auth MA], WC [auth mA], X, Y, YA [auth y], YC [auth oA], ZA [auth z] | 7 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G55220VL GlyCosmos: G55220VL GlyGen: G55220VL | |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | CC [auth SA], G, RA [auth r] | 9 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G36191CD GlyCosmos: G36191CD GlyGen: G36191CD | |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | IC [auth YA], M, XA [auth x] | 8 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G83582BK GlyCosmos: G83582BK GlyGen: G83582BK | |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | FD [auth vA], JA [auth j], UB [auth KA] | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22768VO GlyCosmos: G22768VO GlyGen: G22768VO | |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | JD [auth zA], NA [auth n], YB [auth OA] | 2 |  | O-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 2 |
| MODEL REFINEMENT | PHENIX | 1.19 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01AI148382 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | S10OD021600 |
| Department of Energy (DOE, United States) | United States | -- |
| National Institutes of Health/National Eye Institute (NIH/NEI) | United States | R01GM080139 |
| National Science Foundation (NSF, United States) | United States | MCB-1942763 |
| National Institutes of Health/National Institute of Neurological Disorders and Stroke (NIH/NINDS) | United States | 1R01NS119505-01A1 |
| National Science Foundation (NSF, United States) | United States | ACI-1548562 |
| Department of Energy (DOE, United States) | United States | DE-AC05-00OR22725 |