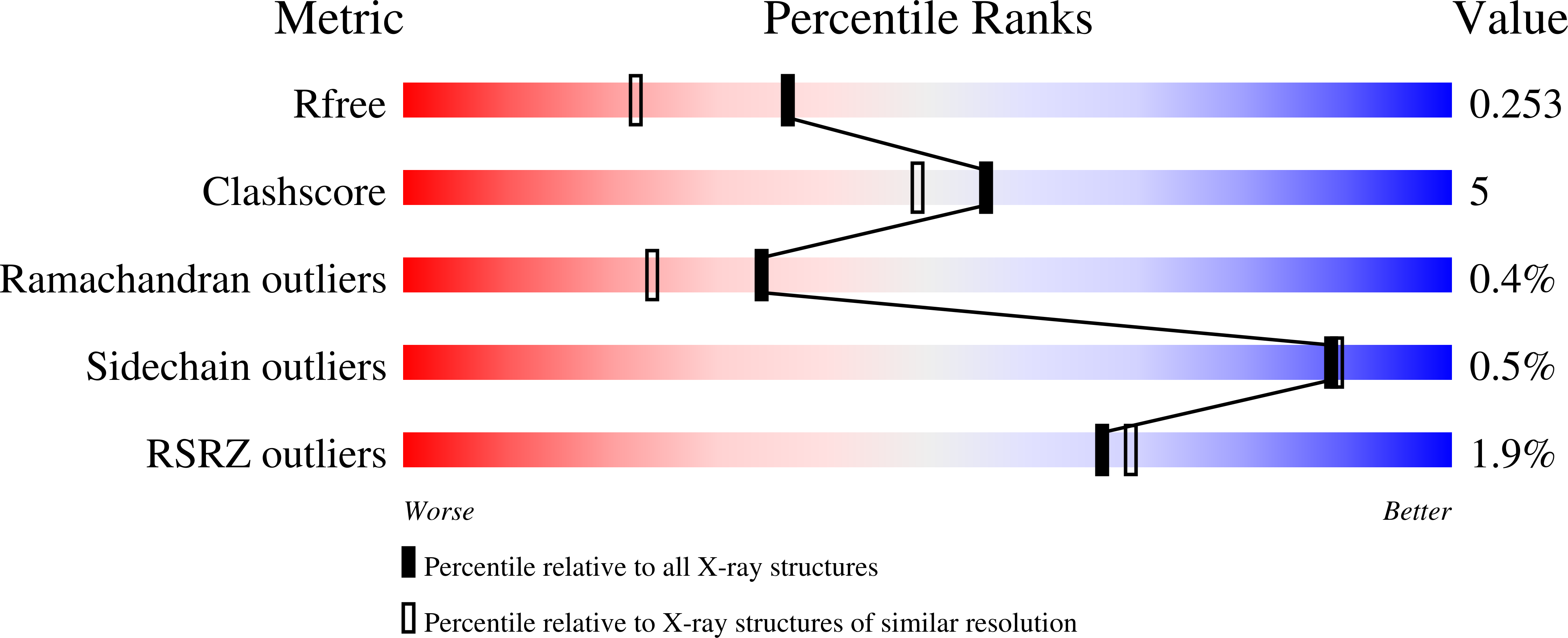
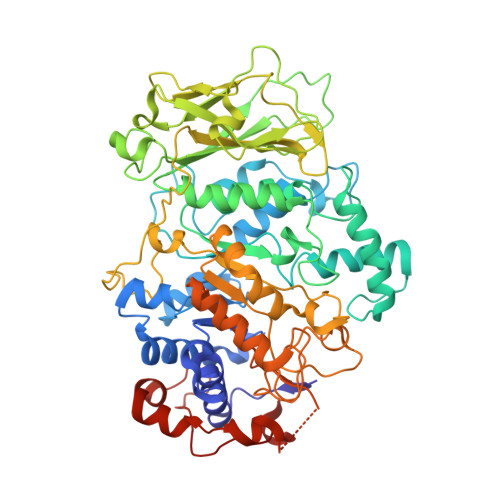
The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis
Schormann, N., Patel, M., Thannickal, L., Purushotham, S., Wu, R., Mieher, J.L., Wu, H., Deivanayagam, C.(2023) Acta Crystallogr F Struct Biol Commun 79: 119-127