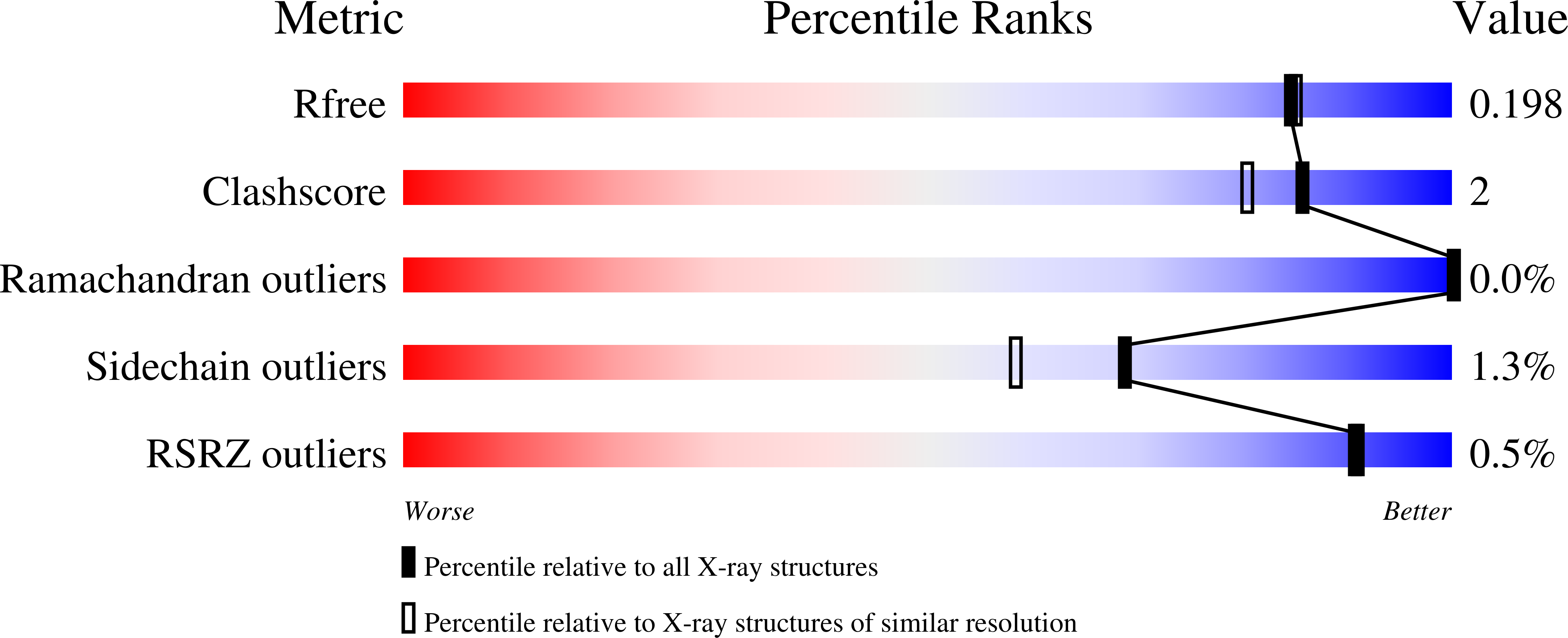
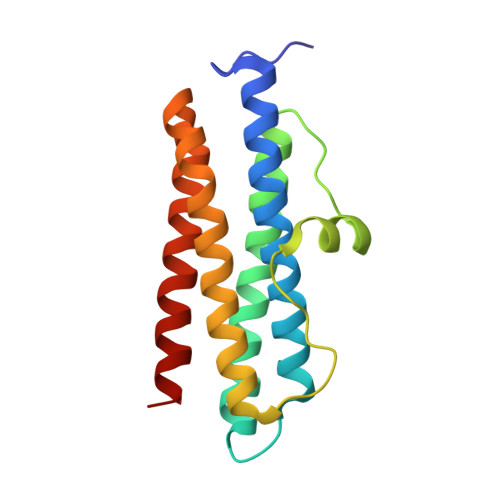
Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Rajapaksha, N., Soldano, A., Yao, H., Donnarumma, F., Kashipathy, M.M., Seibold, S., Battaile, K.P., Lovell, S., Rivera, M.(2023) Int J Mol Sci 24
- PubMed: 36902100
- DOI: https://doi.org/10.3390/ijms24054669
- Primary Citation of Related Structures:
8FF9, 8FFA, 8FFB, 8FFC, 8FFD - PubMed Abstract:
We report the structural, biochemical, and functional characterization of the product of gene PA0962 from Pseudomonas aeruginosa PAO1. The protein, termed Pa Dps, adopts the Dps subunit fold and oligomerizes into a nearly spherical 12-mer quaternary structure at pH 6.0 or in the presence of divalent cations at neutral pH and above. The 12-Mer Pa Dps contains two di-iron centers at the interface of each subunit dimer, coordinated by conserved His, Glu, and Asp residues. In vitro, the di-iron centers catalyze the oxidation of Fe 2+ utilizing H 2 O 2 (not O 2 ) as an oxidant, suggesting Pa Dps functions to aid P. aeruginosa to survive H 2 O 2 -mediated oxidative stress. In agreement, a P. aeruginosa Δ dps mutant is significantly more susceptible to H 2 O 2 than the parent strain. The Pa Dps structure harbors a novel network of Tyr residues at the interface of each subunit dimer between the two di-iron centers, which captures radicals generated during Fe 2+ oxidation at the ferroxidase centers and forms di-tyrosine linkages, thus effectively trapping the radicals within the Dps shell. Surprisingly, incubating Pa Dps and DNA revealed unprecedented DNA cleaving activity that is independent of H 2 O 2 or O 2 but requires divalent cations and 12-mer Pa Dps.
Organizational Affiliation:
Department of Chemistry, Louisiana State University, 232 Choppin Hall, Baton Rouge, LA 70803, USA.