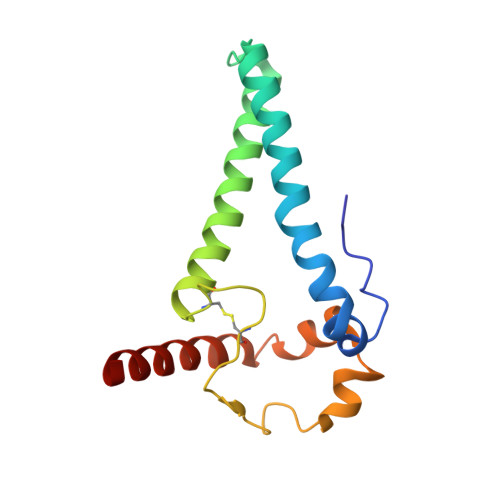
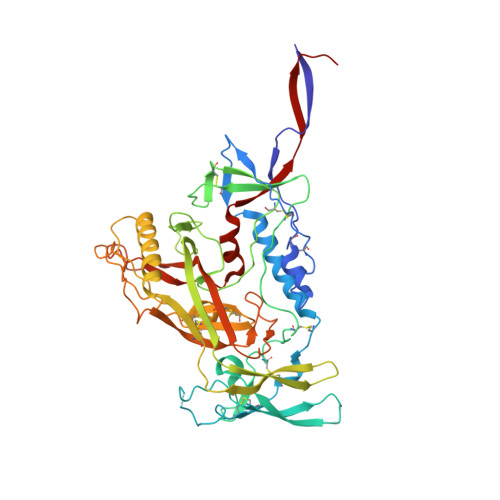
Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles.
Wang, K., Zhang, S., Go, E.P., Ding, H., Wang, W.L., Nguyen, H.T., Kappes, J.C., Desaire, H., Sodroski, J., Mao, Y.(2023) Commun Biol 6: 535-535
- PubMed: 37202420
- DOI: https://doi.org/10.1038/s42003-023-04916-w
- Primary Citation of Related Structures:
8FAD, 8FAE - PubMed Abstract:
During virus entry, the pretriggered human immunodeficiency virus (HIV-1) envelope glycoprotein (Env) trimer initially transits into a default intermediate state (DIS) that remains structurally uncharacterized. Here, we present cryo-EM structures at near-atomic resolution of two cleaved full-length HIV-1 Env trimers purified from cell membranes in styrene-maleic acid lipid nanoparticles without antibodies or receptors. The cleaved Env trimers exhibited tighter subunit packing than uncleaved trimers. Cleaved and uncleaved Env trimers assumed remarkably consistent yet distinct asymmetric conformations, with one smaller and two larger opening angles. Breaking conformational symmetry is allosterically coupled with dynamic helical transformations of the gp41 N-terminal heptad repeat (HR1 N ) regions in two protomers and with trimer tilting in the membrane. The broken symmetry of the DIS potentially assists Env binding to two CD4 receptors-while resisting antibody binding-and promotes extension of the gp41 HR1 helical coiled-coil, which relocates the fusion peptide closer to the target cell membrane.
Organizational Affiliation:
State Key Laboratory for Mesoscopic Physics, School of Physics, Peking University, Beijing, China.