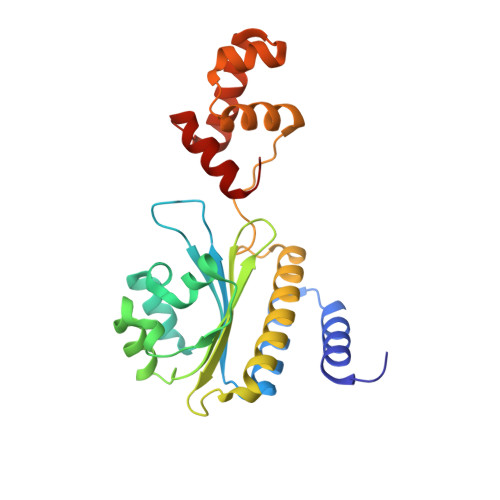
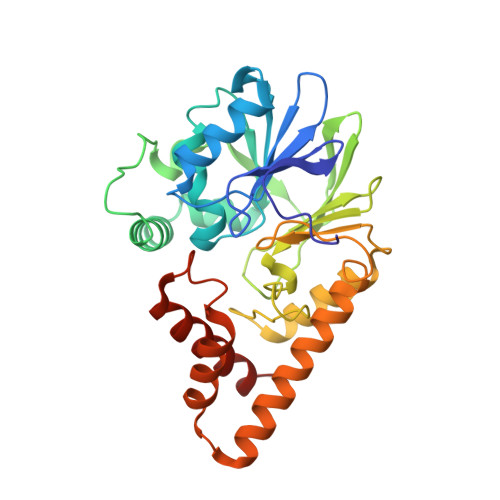
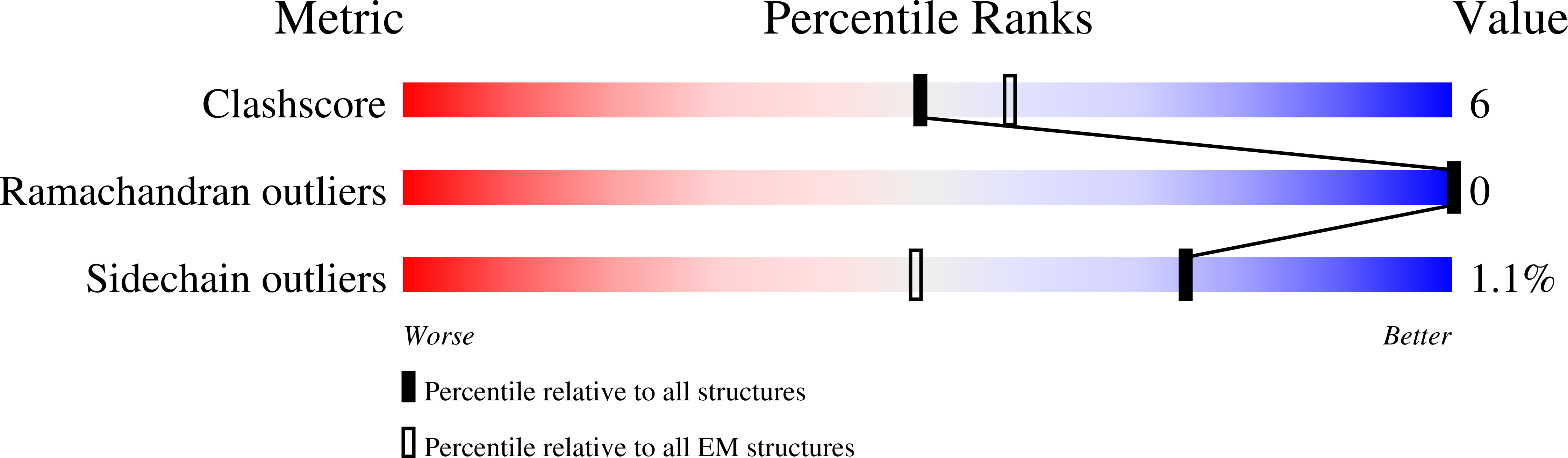Structure of the RhlR-PqsE complex from Pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation.
Feathers, J.R., Richael, E.K., Simanek, K.A., Fromme, J.C., Paczkowski, J.E.(2022) Structure 30: 1626
- PubMed: 36379213
- DOI: https://doi.org/10.1016/j.str.2022.10.008
- Primary Citation of Related Structures:
8DQ0, 8DQ1 - PubMed Abstract:
Pseudomonas aeruginosa is an opportunistic pathogen that is responsible for thousands of deaths every year in the United States. P. aeruginosa virulence factor production is mediated by quorum sensing, a mechanism of bacterial cell-cell communication that relies on the production and detection of signal molecules called autoinducers. In P. aeruginosa, the transcription factor receptor RhlR is activated by a RhlI-synthesized autoinducer. We recently showed that RhlR-dependent transcription is enhanced by a physical interaction with the enzyme PqsE via increased affinity of RhlR for promoter DNA. However, the molecular basis for complex formation and how complex formation enhanced RhlR transcriptional activity remained unclear. Here, we report the structure of ligand-bound RhlR in complex with PqsE. Additionally, we determined the structure of the complex bound with DNA, revealing the mechanism by which RhlR-mediated transcription is enhanced by PqsE, thereby establishing the molecular basis for RhlR-dependent virulence factor production in P. aeruginosa.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Weill Institute for Cell and Molecular Biology, Cornell University, Ithaca, NY 14853, USA.