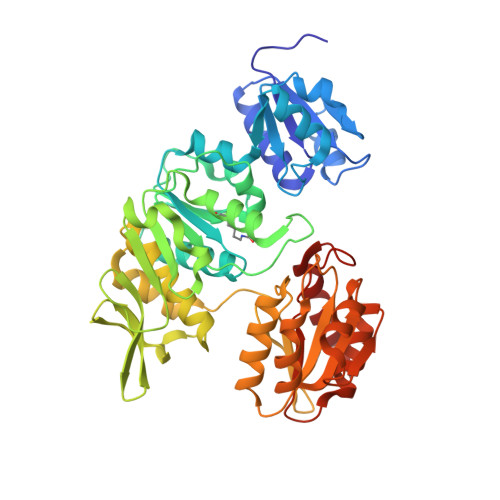
Crystal Structure of UDP-N-acetylmuramoylalanine--D-glutamate ligase (MurD) from Pseudomonas aeruginosa PAO1 in complex with UMA (Uridine-5'-diphosphate-N-acetylmuramoyl-L-Alanine)
Abendroth, J., DeBouver, N.D., Lorimer, D.D., Horanyi, P.S., Edwards, T.E.To be published.