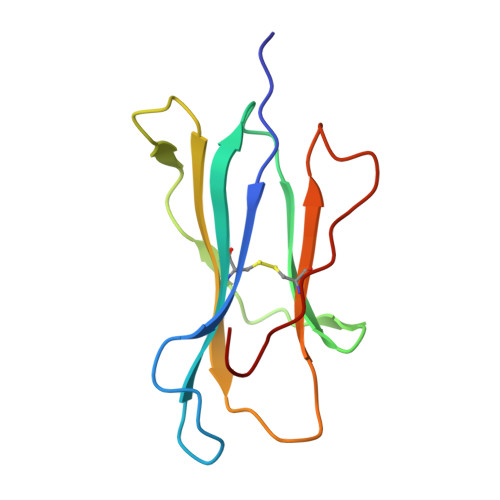
SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Choy, C., Chen, J., Li, J., Gallagher, D.T., Lu, J., Wu, D., Zou, A., Hemani, H., Baptiste, B.A., Wichmann, E., Yang, Q., Ciffelo, J., Yin, R., McKelvy, J., Melvin, D., Wallace, T., Dunn, C., Nguyen, C., Chia, C.W., Fan, J., Ruffolo, J., Zukley, L., Shi, G., Amano, T., An, Y., Meirelles, O., Wu, W.W., Chou, C.K., Shen, R.F., Willis, R.A., Ko, M.S.H., Liu, Y.T., De, S., Pierce, B.G., Ferrucci, L., Egan, J., Mariuzza, R., Weng, N.P.(2023) Nat Commun 14: 6725-6725
- PubMed: 37872153
- DOI: https://doi.org/10.1038/s41467-023-42430-z
- Primary Citation of Related Structures:
8DNT - PubMed Abstract:
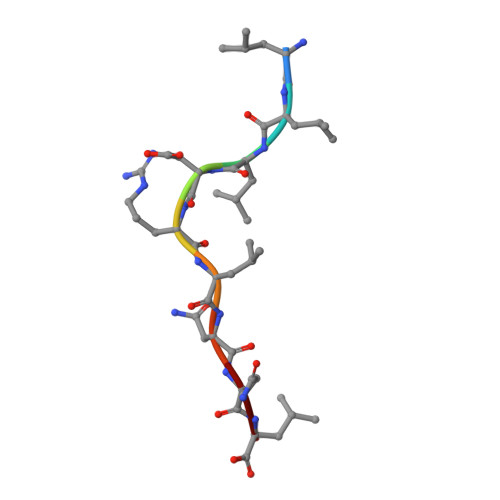
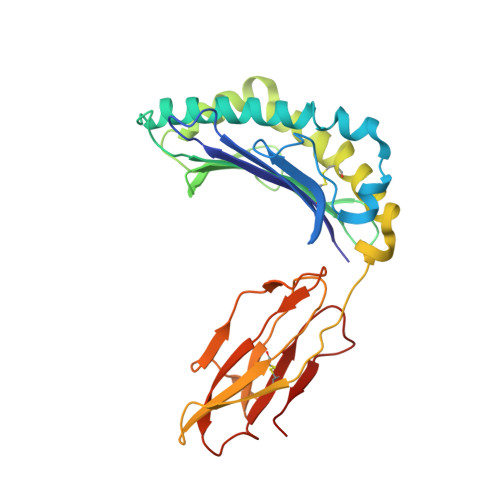
The resolution of SARS-CoV-2 replication hinges on cell-mediated immunity, wherein CD8 + T cells play a vital role. Nonetheless, the characterization of the specificity and TCR composition of CD8 + T cells targeting non-spike protein of SARS-CoV-2 before and after infection remains incomplete. Here, we analyzed CD8 + T cells recognizing six epitopes from the SARS-CoV-2 nucleocapsid (N) protein and found that SARS-CoV-2 infection slightly increased the frequencies of N-recognizing CD8 + T cells but significantly enhanced activation-induced proliferation compared to that of the uninfected donors. The frequencies of N-specific CD8 + T cells and their proliferative response to stimulation did not decrease over one year. We identified the N 222-230 peptide (LLLDRLNQL, referred to as LLL thereafter) as a dominant epitope that elicited the greatest proliferative response from both convalescent and uninfected donors. Single-cell sequencing of T cell receptors (TCR) from LLL-specific CD8 + T cells revealed highly restricted Vα gene usage (TRAV12-2) with limited CDR3α motifs, supported by structural characterization of the TCR-LLL-HLA-A2 complex. Lastly, transcriptome analysis of LLL-specific CD8 + T cells from donors who had expansion (expanders) or no expansion (non-expanders) after in vitro stimulation identified increased chromatin modification and innate immune functions of CD8 + T cells in non-expanders. These results suggests that SARS-CoV-2 infection induces LLL-specific CD8 + T cell responses with a restricted TCR repertoire.
Organizational Affiliation:
Laboratory of Molecular Biology and Immunology, National Institute on Aging, NIH, Baltimore, MD, USA.