Cryo-EM Structure and Functional Studies of EBNA1 Binding to the Family of Repeats and Dyad Symmetry Elements of Epstein-Barr Virus oriP.
Mei, Y., Messick, T.E., Dheekollu, J., Kim, H.J., Molugu, S., Munoz, L.J.C., Moiskeenkova-Bell, V., Murakami, K., Lieberman, P.M.(2022) J Virol 96: e0094922-e0094922
- PubMed: 36037477
- DOI: https://doi.org/10.1128/jvi.00949-22
- Primary Citation of Related Structures:
8DLF - PubMed Abstract:
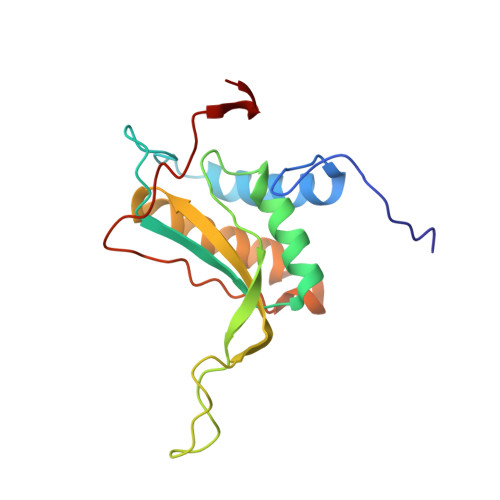
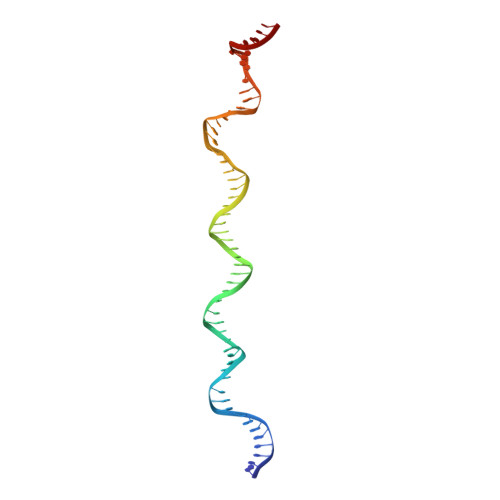
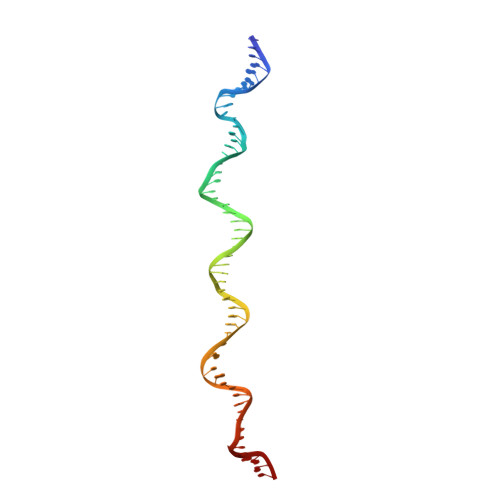
Epstein-Barr nuclear antigen 1 (EBNA1) is a multifunctional viral-encoded DNA-binding protein essential for Epstein-Barr virus (EBV) DNA replication and episome maintenance. EBNA1 binds to two functionally distinct elements at the viral origin of plasmid replication ( oriP ), termed the dyad symmetry (DS) element, required for replication initiation and the family of repeats (FR) required for episome maintenance. Here, we determined the cryo-electron microscopy (cryo-EM) structure of the EBNA1 DNA binding domain (DBD) from amino acids (aa) 459 to 614 and its interaction with two tandem sites at the DS and FR. We found that EBNA1 induces a strong DNA bending angle in the DS, while the FR is more linear. The N-terminal arm of the DBD (aa 444 to 468) makes extensive contact with DNA as it wraps around the minor groove, with some conformational variation among EBNA1 monomers. Mutation of variable-contact residues K460 and K461 had only minor effects on DNA binding but had abrogated oriP -dependent DNA replication. We also observed that the AT-rich intervening DNA between EBNA1 binding sites in the FR can be occupied by the EBNA1 AT hook, N-terminal domain (NTD) aa 1 to 90 to form a Zn-dependent stable complex with EBNA1 DBD on a 2×FR DNA template. We propose a model showing EBNA1 DBD and NTD cobinding at the FR and suggest that this may contribute to the oligomerization of viral episomes important for maintenance during latent infection. IMPORTANCE EBV latent infection is causally linked to diverse cancers and autoimmune disorders. EBNA1 is the viral-encoded DNA binding protein required for episomal maintenance during latent infection and is consistently expressed in all EBV tumors. The interaction of EBNA1 with different genetic elements confers different viral functions, such as replication initiation at DS and chromosome tethering at FR. Here, we used cryo-EM to determine the structure of the EBNA1 DNA-binding domain (DBD) bound to two tandem sites at the DS and at the FR. We also show that the NTD of EBNA1 can interact with the AT-rich DNA sequence between tandem EBNA1 DBD binding sites in the FR. These results provide new information on the mechanism of EBNA1 DNA binding at DS and FR and suggest a higher-order oligomeric structure of EBNA1 bound to FR. Our findings have implications for targeting EBNA1 in EBV-associated disease.
Organizational Affiliation:
The Wistar Institute, Philadelphia, Pennsylvania, USA.