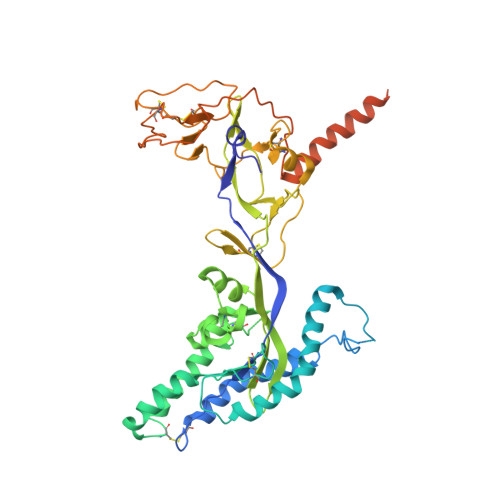
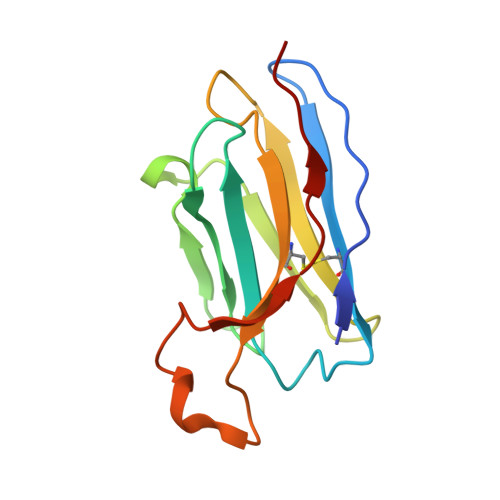
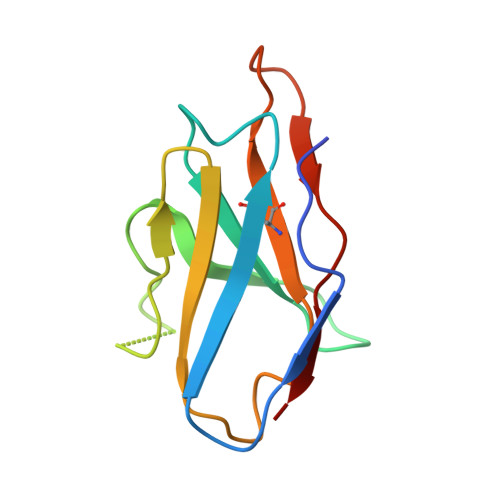
Cross-protective antibodies against common endemic respiratory viruses.
Caban, M., Rodarte, J.V., Bibby, M., Gray, M.D., Taylor, J.J., Pancera, M., Boonyaratanakornkit, J.(2023) Nat Commun 14: 798-798
- PubMed: 36781872
- DOI: https://doi.org/10.1038/s41467-023-36459-3
- Primary Citation of Related Structures:
8DG8, 8DG9 - PubMed Abstract:
Respiratory syncytial virus (RSV), human metapneumovirus (HMPV), and human parainfluenza virus types one (HPIV1) and three (HPIV3) can cause severe disease and death in immunocompromised patients, the elderly, and those with underlying lung disease. A protective monoclonal antibody exists for RSV, but clinical use is limited to high-risk infant populations. Hence, therapeutic options for these viruses in vulnerable patient populations are currently limited. Here, we present the discovery, in vitro characterization, and in vivo efficacy testing of two cross-neutralizing monoclonal antibodies, one targeting both HPIV3 and HPIV1 and the other targeting both RSV and HMPV. The 3 × 1 antibody is capable of targeting multiple parainfluenza viruses; the MxR antibody shares features with other previously reported monoclonal antibodies that are capable of neutralizing both RSV and HMPV. We obtained structures using cryo-electron microscopy of these antibodies in complex with their antigens at 3.62 Å resolution for 3 × 1 bound to HPIV3 and at 2.24 Å for MxR bound to RSV, providing a structural basis for in vitro binding and neutralization. Together, a cocktail of 3 × 1 and MxR could have clinical utility in providing broad protection against four of the respiratory viruses that cause significant morbidity and mortality in at-risk individuals.
Organizational Affiliation:
Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Center, Seattle, WA, USA.