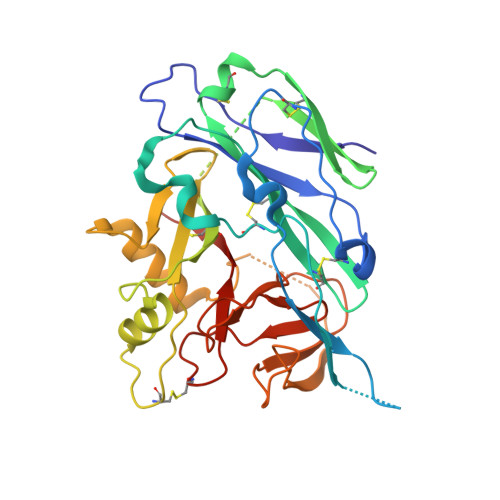
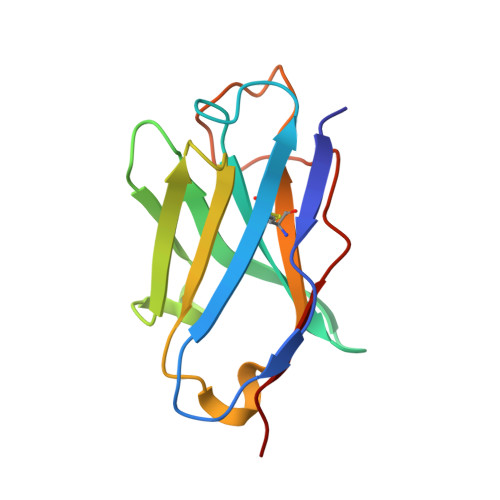
Antigenic mapping and functional characterization of human new world hantavirus neutralizing antibodies.
Engdahl, T.B., Binshtein, E., Brocato, R.L., Kuzmina, N.A., Principe, L.M., Kwilas, S.A., Kim, R.K., Chapman, N.S., Porter, M.S., Guardado-Calvo, P., Rey, F.A., Handal, L.S., Diaz, S.M., Zagol-Ikapitte, I.A., Tran, M.H., McDonald, W.H., Meiler, J., Reidy, J.X., Trivette, A., Bukreyev, A., Hooper, J.W., Crowe Jr., J.E.(2023) Elife 12
- PubMed: 36971354
- DOI: https://doi.org/10.7554/eLife.81743
- Primary Citation of Related Structures:
8DBZ - PubMed Abstract:
Hantaviruses are high-priority emerging pathogens carried by rodents and transmitted to humans by aerosolized excreta or, in rare cases, person-to-person contact. While infections in humans are relatively rare, mortality rates range from 1 to 40% depending on the hantavirus species. There are currently no FDA-approved vaccines or therapeutics for hantaviruses, and the only treatment for infection is supportive care for respiratory or kidney failure. Additionally, the human humoral immune response to hantavirus infection is incompletely understood, especially the location of major antigenic sites on the viral glycoproteins and conserved neutralizing epitopes. Here, we report antigenic mapping and functional characterization for four neutralizing hantavirus antibodies. The broadly neutralizing antibody SNV-53 targets an interface between Gn/Gc, neutralizes through fusion inhibition and cross-protects against the Old World hantavirus species Hantaan virus when administered pre- or post-exposure. Another broad antibody, SNV-24, also neutralizes through fusion inhibition but targets domain I of Gc and demonstrates weak neutralizing activity to authentic hantaviruses. ANDV-specific, neutralizing antibodies (ANDV-5 and ANDV-34) neutralize through attachment blocking and protect against hantavirus cardiopulmonary syndrome (HCPS) in animals but target two different antigenic faces on the head domain of Gn. Determining the antigenic sites for neutralizing antibodies will contribute to further therapeutic development for hantavirus-related diseases and inform the design of new broadly protective hantavirus vaccines.
Organizational Affiliation:
Department of Pathology, Microbiology and Immunology, Vanderbilt University, Nashville, United States.