Recognition and Cleavage of Human tRNA Methyltransferase TRMT1 by the SARS-CoV-2 Main Protease.
D'Oliviera, A., Dai, X., Mottaghinia, S., Geissler, E.P., Etienne, L., Zhang, Y., Mugridge, J.S.(2023) Elife
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2023) Elife
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 3C-like proteinase nsp5 | 308 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 1 Gene Names: rep, 1a-1b EC: 3.4.22.69 | 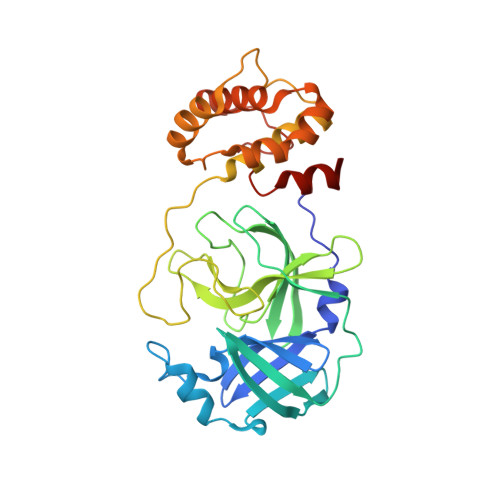 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| tRNA (guanine(26)-N(2))-dimethyltransferase | 11 | Homo sapiens | Mutation(s): 0 EC: 2.1.1.216 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9NXH9 (Homo sapiens) Explore Q9NXH9 Go to UniProtKB: Q9NXH9 | |||||
PHAROS: Q9NXH9 GTEx: ENSG00000104907 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9NXH9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | F [auth A], H [auth B], I [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CL Query on CL | D [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | E [auth A], G [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 67.787 | α = 90 |
| b = 100.029 | β = 90 |
| c = 103.252 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM143000 |