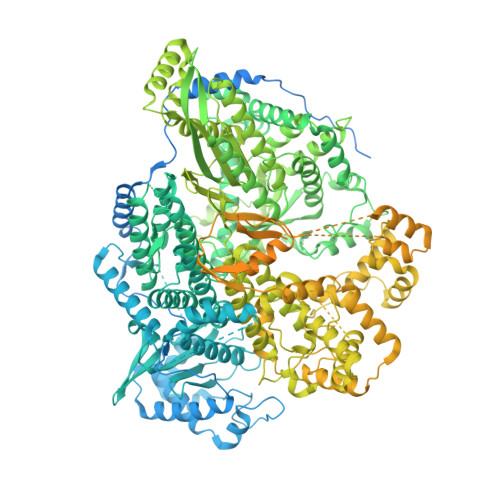
Structures of active Hantaan virus polymerase uncover the mechanisms of Hantaviridae genome replication.
Durieux Trouilleton, Q., Barata-Garcia, S., Arragain, B., Reguera, J., Malet, H.(2023) Nat Commun 14: 2954-2954
- PubMed: 37221161
- DOI: https://doi.org/10.1038/s41467-023-38555-w
- Primary Citation of Related Structures:
8C4S, 8C4T, 8C4U, 8C4V - PubMed Abstract:
Hantaviruses are causing life-threatening zoonotic infections in humans. Their tripartite negative-stranded RNA genome is replicated by the multi-functional viral RNA-dependent RNA-polymerase. Here we describe the structure of the Hantaan virus polymerase core and establish conditions for in vitro replication activity. The apo structure adopts an inactive conformation that involves substantial folding rearrangement of polymerase motifs. Binding of the 5' viral RNA promoter triggers Hantaan virus polymerase reorganization and activation. It induces the recruitment of the 3' viral RNA towards the polymerase active site for prime-and-realign initiation. The elongation structure reveals the formation of a template/product duplex in the active site cavity concomitant with polymerase core widening and the opening of a 3' viral RNA secondary binding site. Altogether, these elements reveal the molecular specificities of Hantaviridae polymerase structure and uncover the mechanisms underlying replication. They provide a solid framework for future development of antivirals against this group of emerging pathogens.
Organizational Affiliation:
Univ. Grenoble Alpes, CNRS, CEA, IBS, F-38000, Grenoble, France.