Identification of New L-Fucosyl and L-Galactosyl Amides as Glycomimetic Ligands of TNF Lectin Domain of BC2L-C from Burkholderia cenocepacia.
Mazzotta, S., Antonini, G., Vasile, F., Gillon, E., Lundstrom, J., Varrot, A., Belvisi, L., Bernardi, A.(2023) Molecules 28
- PubMed: 36771163
- DOI: https://doi.org/10.3390/molecules28031494
- Primary Citation of Related Structures:
8BRO - PubMed Abstract:
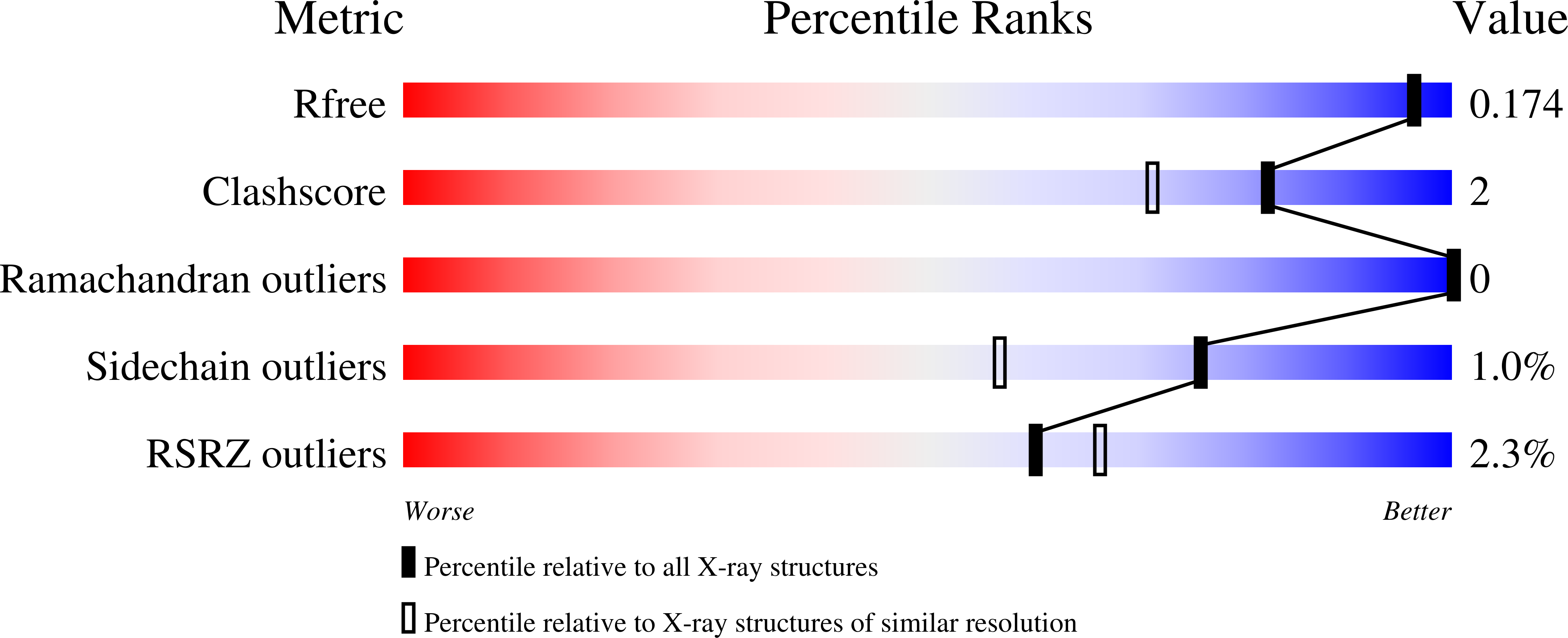
The inhibition of carbohydrate-lectin interactions is being explored as an efficient approach to anti adhesion therapy and biofilm destabilization, two alternative antimicrobial strategies that are being explored against resistant pathogens. BC2L-C is a new type of lectin from Burkholderia cenocepacia that binds (mammalian) fucosides at the N -terminal domain and (bacterial) mannosides at the C -terminal domain. This double carbohydrate specificity allows the lectin to crosslink host cells and bacterial cells. We have recently reported the design and generation of the first glycomimetic antagonists of BC2L-C, β- C - or β- N -fucosides that target the fucose-specific N- terminal domain (BC2L-C-Nt). The low water solubility of the designed N -fucosides prevented a full examination of this promising series of ligands. In this work, we describe the synthesis and biophysical evaluation of new L-fucosyl and L-galactosyl amides, designed to be water soluble and to interact with BC2L-C-Nt. The protein-ligand interaction was investigated by Saturation Transfer Difference NMR, Isothermal Titration Calorimetry and crystallographic studies. STD-NMR experiments showed that both fucosyl and galactosyl amides compete with α-methyl fucoside for lectin binding. A new hit compound was identified with good water solubility and an affinity for BC2L-C-Nt of 159 μM (ITC), which represents a one order of magnitude gain over α-methyl fucoside. The x-ray structure of its complex with BC2L-C-Nt was solved at 1.55 Å resolution.
Organizational Affiliation:
Dipartimento di Chimica, Università degli Studi di Milano, 20133 Milan, Italy.