Dissection of a Native Bacterial Xyloglucan-degrading System using Activity-Based Probes
McGregor, N.G.S., de Boer, C., Foucart, Q., Beenakker, T., Willems, L., Overkleeft, H.S., Davies, G.J.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cellulase, putative, cel5C | 346 | Cellvibrio japonicus Ueda107 | Mutation(s): 0 Gene Names: cel5C | 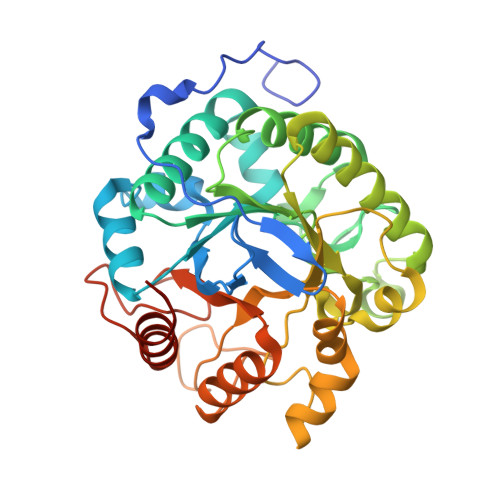 | |
UniProt | |||||
Find proteins for B3PF55 (Cellvibrio japonicus (strain Ueda107)) Explore B3PF55 Go to UniProtKB: B3PF55 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B3PF55 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| YLL (Subject of Investigation/LOI) Query on YLL | D [auth A], F [auth B], I [auth C] | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL C7 H14 O6 QFYQIWDMMSKNFF-NYLBLOMBSA-N |  | ||
| BGC (Subject of Investigation/LOI) Query on BGC | E [auth A], G [auth B], H [auth C] | beta-D-glucopyranose C6 H12 O6 WQZGKKKJIJFFOK-VFUOTHLCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 126.441 | α = 90 |
| b = 176.159 | β = 90 |
| c = 115.701 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| autoPROC | data reduction |
| autoPROC | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/R001162/1 |
| European Research Council (ERC) | European Union | 2020-SyG-951231 |