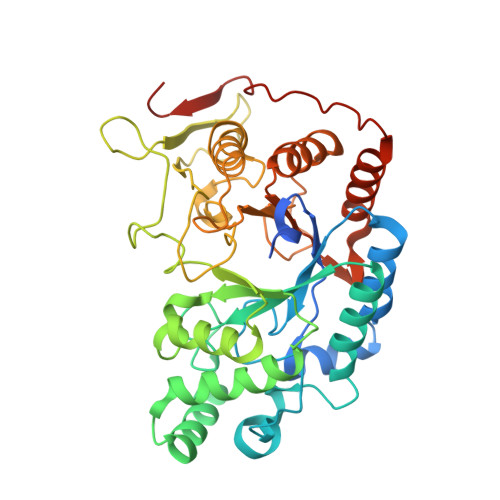
Structural and functional characterization of the novel endo-alpha (1,4)-fucoidanase Mef1 from the marine bacterium Muricauda eckloniae.
Mikkelsen, M.D., Tran, V.H.N., Meier, S., Nguyen, T.T., Holck, J., Cao, H.T.T., Van, T.T.T., Thinh, P.D., Meyer, A.S., Morth, J.P.(2023) Acta Crystallogr D Struct Biol 79: 1026-1043
- PubMed: 37877949
- DOI: https://doi.org/10.1107/S2059798323008732
- Primary Citation of Related Structures:
8BPD - PubMed Abstract:
Fucoidanases (EC 3.2.1.-) catalyze the hydrolysis of glycosidic bonds between fucose residues in fucoidans. Fucoidans are a compositionally and structurally diverse class of fucose-containing sulfated polysaccharides that are primarily found in brown seaweeds. Here, the structural characterization of a novel endo-α(1,4)-fucoidanase, Mef1, from the marine bacterium Muricauda eckloniae is presented, showing sequence similarity to members of glycoside hydrolase family 107. Using carbohydrate polyacrylamide gel electrophoresis and nuclear magnetic resonance analyses, it is shown that the fucoidanase Mef1 catalyzes the cleavage of α(1,4)-linkages between fucose residues sulfated on C2 in the structure [-3)-α-L-Fucp2S-(1,4)-α-L-Fucp2S-(1-] n in fucoidan from Fucus evanescens. Kinetic analysis of Mef1 activity by Fourier transform infrared spectroscopy revealed that the specific Mef1 fucoidanase activity (U f ) on F. evanescens fucoidan was 0.1 × 10 -3 U f µM -1 . By crystal structure determination of Mef1 at 1.8 Å resolution, a single-domain organization comprising a (β/α) 8 -barrel domain was determined. The active site was in an extended, positively charged groove that is likely to be designed to accommodate the binding of the negatively charged, sulfated fucoidan substrate. The active site of Mef1 comprises the amino acids His270 and Asp187, providing acid/base and nucleophile groups, respectively, for the hydrolysis of glycosidic bonds in the fucoidan backbone. Electron densities were identified for two possible Ca 2+ ions in the enzyme, one of which is partially exposed to the active-site groove, while the other is very tightly coordinated. A water wire was discovered leading from the exterior of the Mef1 enzyme into the active site, passing the tightly coordinated Ca 2+ site.
Organizational Affiliation:
Protein Chemistry and Enzyme Technology Section, Department of Biotechnology and Biomedicine, Technical University of Denmark, DK-2800 Kgs Lyngby, Denmark.