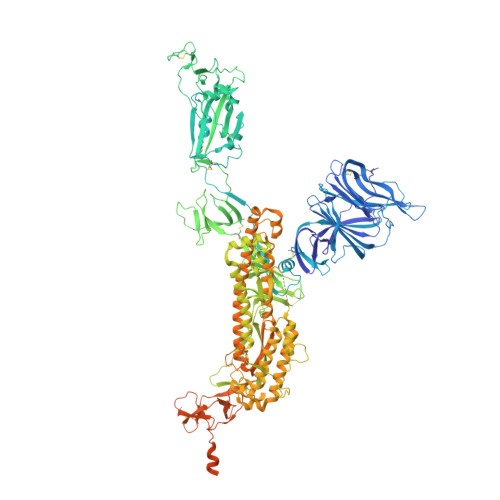
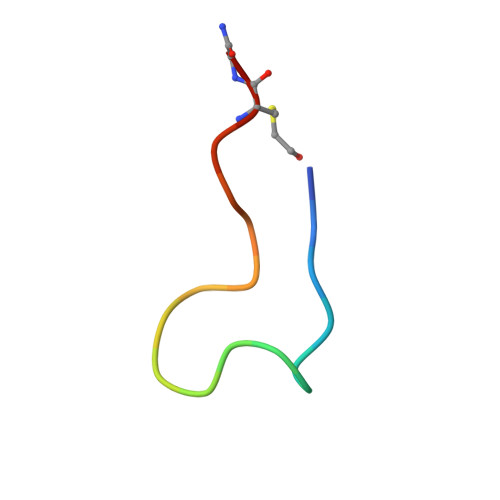
A broad-spectrum macrocyclic peptide inhibitor of the SARS-CoV-2 spike protein.
Thijssen, V., Hurdiss, D.L., Debski-Antoniak, O.J., Spence, M.A., Franck, C., Norman, A., Aggarwal, A., Mokiem, N.J., van Dongen, D.A.A., Vermeir, S.W., Liu, M., Li, W., Chatziandreou, M., Donselaar, T., Du, W., Drulyte, I., Bosch, B.J., Snijder, J., Turville, S.G., Payne, R.J., Jackson, C.J., van Kuppeveld, F.J.M., Jongkees, S.A.K.(2023) Proc Natl Acad Sci U S A 120: e2303292120-e2303292120
- PubMed: 37339194
- DOI: https://doi.org/10.1073/pnas.2303292120
- Primary Citation of Related Structures:
8BON - PubMed Abstract:
The ongoing COVID-19 pandemic has had great societal and health consequences. Despite the availability of vaccines, infection rates remain high due to immune evasive Omicron sublineages. Broad-spectrum antivirals are needed to safeguard against emerging variants and future pandemics. We used messenger RNA (mRNA) display under a reprogrammed genetic code to find a spike-targeting macrocyclic peptide that inhibits SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) Wuhan strain infection and pseudoviruses containing spike proteins of SARS-CoV-2 variants or related sarbecoviruses. Structural and bioinformatic analyses reveal a conserved binding pocket between the receptor-binding domain, N-terminal domain, and S2 region, distal to the angiotensin-converting enzyme 2 receptor-interaction site. Our data reveal a hitherto unexplored site of vulnerability in sarbecoviruses that peptides and potentially other drug-like molecules can target.
Organizational Affiliation:
Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Utrecht University, Utrecht 3584 CG, the Netherlands.