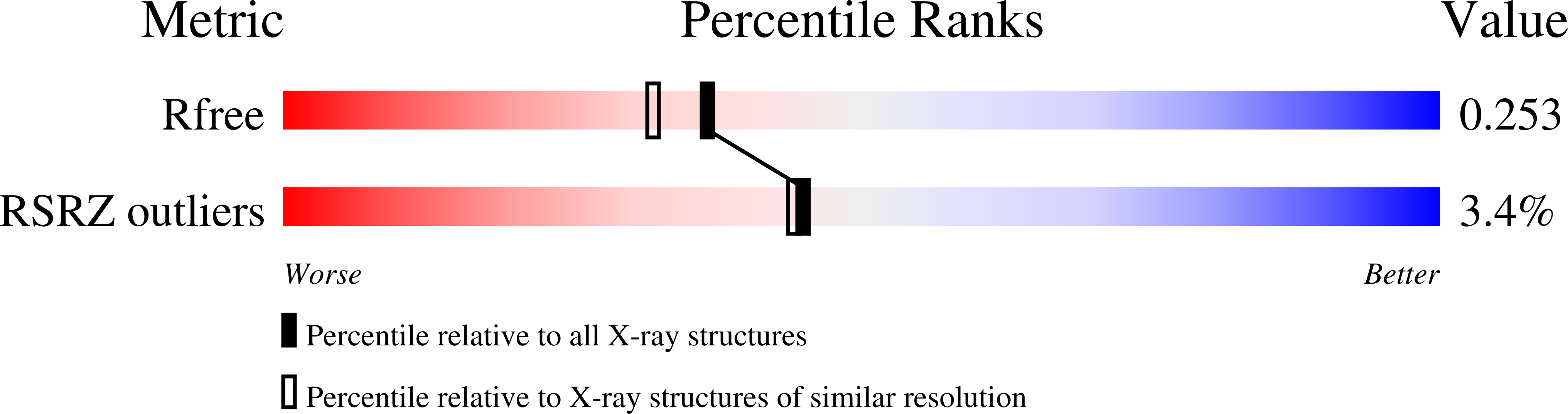Klebsiella phage KP34gp57 capsular depolymerase structure and function: from a serendipitous finding to the design of active mini-enzymes against K. pneumoniae.
Maciejewska, B., Squeglia, F., Latka, A., Privitera, M., Olejniczak, S., Switala, P., Ruggiero, A., Marasco, D., Kramarska, E., Drulis-Kawa, Z., Berisio, R.(2023) mBio 14: e0132923-e0132923
- PubMed: 37707438
- DOI: https://doi.org/10.1128/mbio.01329-23
- Primary Citation of Related Structures:
8BKE - PubMed Abstract:
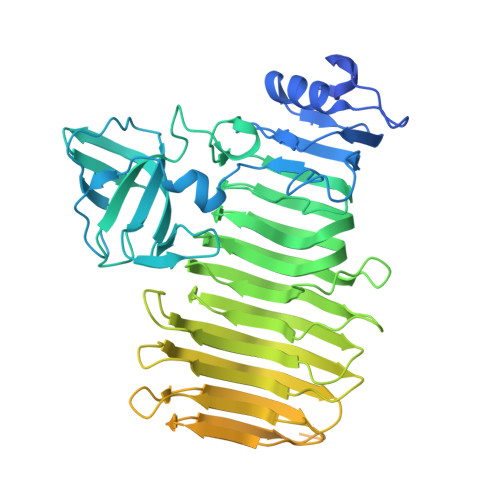
In this work, we determined the structure of Klebsiella phage KP34p57 capsular depolymerase and dissected the role of individual domains in trimerization and functional activity. The crystal structure serendipitously revealed that the enzyme can exist in a monomeric state once deprived of its C-terminal domain. Based on the crystal structure and site-directed mutagenesis, we localized the key catalytic residues in an intra-subunit deep groove. Consistently, we show that C-terminally trimmed KP34p57 variants are monomeric, stable, and fully active. The elaboration of monomeric, fully active phage depolymerases is innovative in the field, as no previous example exists. Indeed, mini phage depolymerases can be combined in chimeric enzymes to extend their activity ranges, allowing their use against multiple serotypes.
Organizational Affiliation:
Department of Pathogen Biology and Immunology, University of Wrocław , Wrocław, Poland.