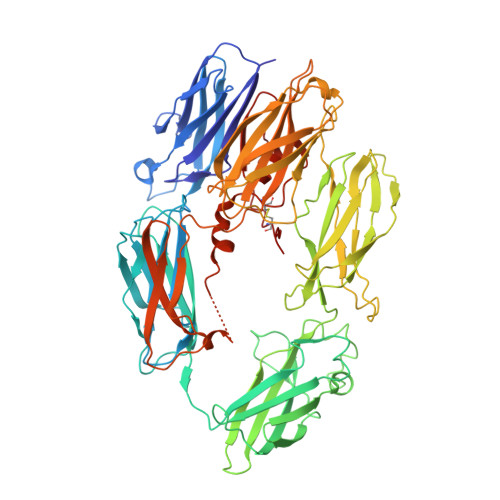
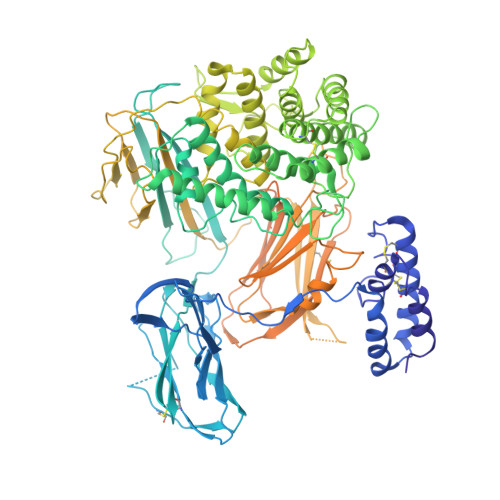
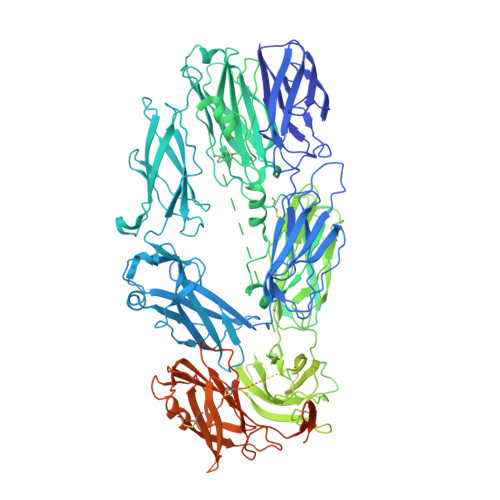
A small-molecule inhibitor of C5 complement protein.
Jendza, K., Kato, M., Salcius, M., Srinivas, H., De Erkenez, A., Nguyen, A., McLaughlin, D., Be, C., Wiesmann, C., Murphy, J., Bolduc, P., Mogi, M., Duca, J., Namil, A., Capparelli, M., Darsigny, V., Meredith, E., Tichkule, R., Ferrara, L., Heyder, J., Liu, F., Horton, P.A., Romanowski, M.J., Schirle, M., Mainolfi, N., Anderson, K., Michaud, G.A.(2019) Nat Chem Biol 15: 666-668
- PubMed: 31209353
- DOI: https://doi.org/10.1038/s41589-019-0303-9
- Primary Citation of Related Structures:
8AYH - PubMed Abstract:
The complement pathway is an important part of the immune system, and uncontrolled activation is implicated in many diseases. The human complement component 5 protein (C5) is a validated drug target within the complement pathway, as an anti-C5 antibody (Soliris) is an approved therapy for paroxysmal nocturnal hemoglobinuria. Here, we report the identification, optimization and mechanism of action for the first small-molecule inhibitor of C5 complement protein.
Organizational Affiliation:
Global Discovery Chemistry, Novartis Institutes of Biomedical Research, Cambridge, MA, USA.