Enzymatic Late-Stage Halogenation of Peptides.
Schnepel, C., Moritzer, A.C., Gafe, S., Montua, N., Minges, H., Niess, A., Niemann, H.H., Sewald, N.(2023) Chembiochem 24: e202200569-e202200569
- PubMed: 36259362
- DOI: https://doi.org/10.1002/cbic.202200569
- Primary Citation of Related Structures:
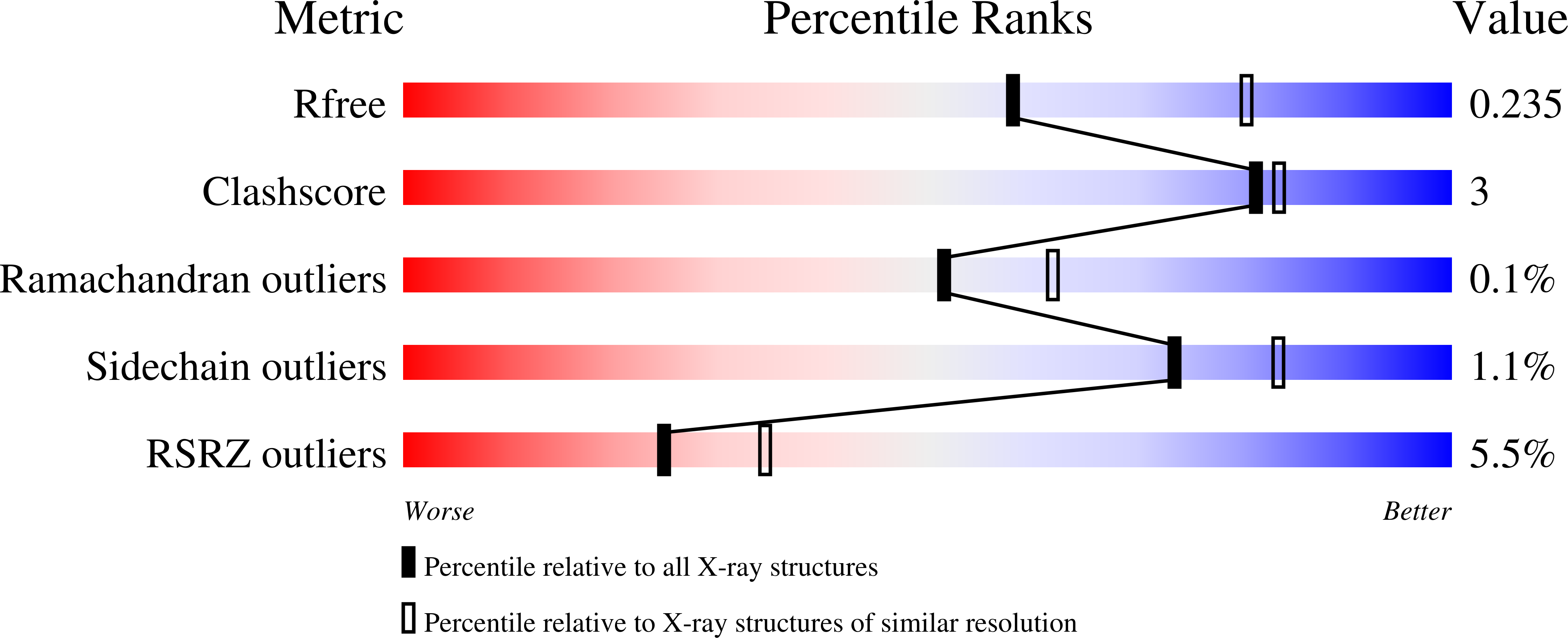
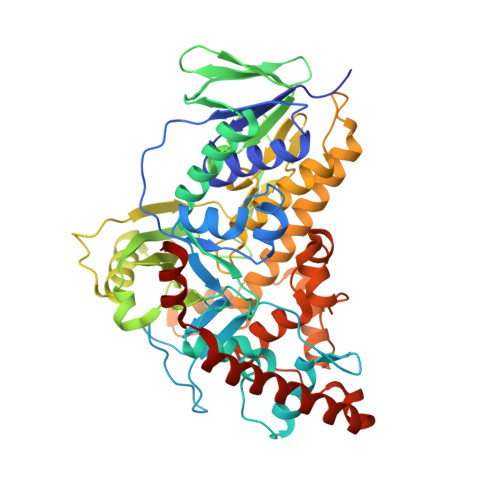
8AD7, 8AD8 - PubMed Abstract:
The late-stage site-selective derivatisation of peptides has many potential applications in structure-activity relationship studies and postsynthetic modification or conjugation of bioactive compounds. The development of orthogonal methods for C-H functionalisation is crucial for such peptide derivatisation. Among them, biocatalytic methods are increasingly attracting attention. Tryptophan halogenases emerged as valuable catalysts to functionalise tryptophan (Trp), while direct enzyme-catalysed halogenation of synthetic peptides is yet unprecedented. Here, it is reported that the Trp 6-halogenase Thal accepts a wide range of amides and peptides containing a Trp moiety. Increasing the sequence length and reaction optimisation made bromination of pentapeptides feasible with good turnovers and a broad sequence scope, while regioselectivity turned out to be sequence dependent. Comparison of X-ray single crystal structures of Thal in complex with d-Trp and a dipeptide revealed a significantly altered binding mode for the peptide. The viability of this bioorthogonal approach was exemplified by halogenation of a cyclic RGD peptide.
Organizational Affiliation:
Organische und Bioorganische Chemie, Fakultät für Chemie, Universität Bielefeld, Universitätsstraße 25, 33615, Bielefeld, Germany.