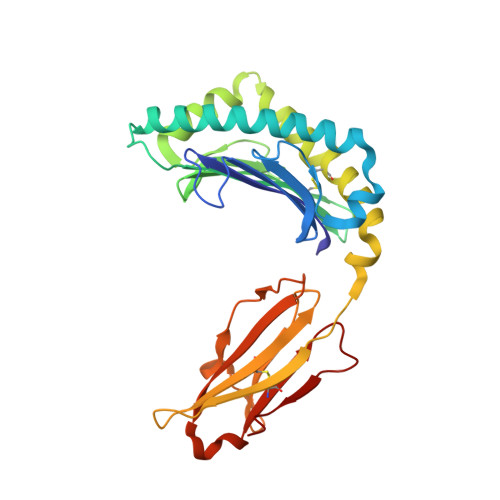
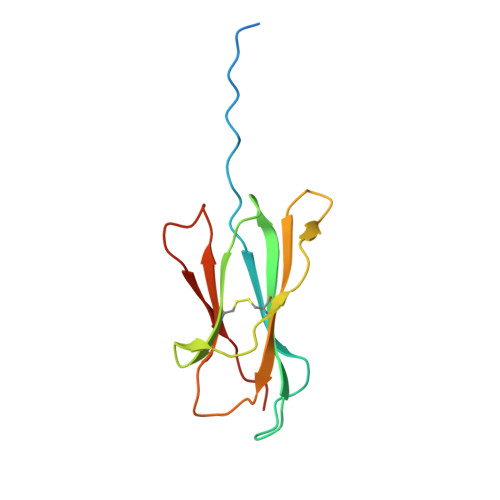
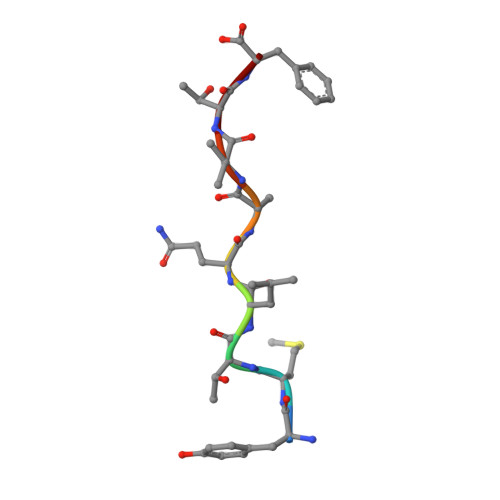
The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Eltschkner, S., Mellinger, S., Buus, S., Nielsen, M., Paulsson, K.M., Lindkvist-Petersson, K., Westerdahl, H.(2023) Front Immunol 14: 1209059-1209059
- PubMed: 37483599
- DOI: https://doi.org/10.3389/fimmu.2023.1209059
- Primary Citation of Related Structures:
7ZQI, 7ZQJ - PubMed Abstract:
Long-distance migratory animals such as birds and bats have evolved to withstand selection imposed by pathogens across the globe, and pathogen richness is known to be particularly high in tropical regions. Immune genes, so-called Major Histocompatibility Complex (MHC) genes, are highly duplicated in songbirds compared to other vertebrates, and this high MHC diversity has been hypothesised to result in a unique adaptive immunity. To understand the rationale behind the evolution of the high MHC genetic diversity in songbirds, we determined the structural properties of an MHC class I protein, Acar3, from a long-distance migratory songbird, the great reed warbler Acrocephalus arundinaceus (in short: Acar ). The structure of Acar3 was studied in complex with pathogen-derived antigens and shows an overall antigen presentation similar to human MHC class I. However, the peptides bound to Acar3 display an unusual conformation: Whereas the N-terminal ends of the peptides display enhanced flexibility, the conformation of their C-terminal halves is rather static. This uncommon peptide-binding mode in Acar3 is facilitated by a central Arg residue within the peptide-binding groove that fixes the backbone of the peptide at its central position, and potentially permits successful interactions between MHC class I and innate immune receptors. Our study highlights the importance of investigating the immune system of wild animals, such as birds and bats, to uncover unique immune mechanisms which may neither exist in humans nor in model organisms.
Organizational Affiliation:
Molecular Plant Pathology, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands.