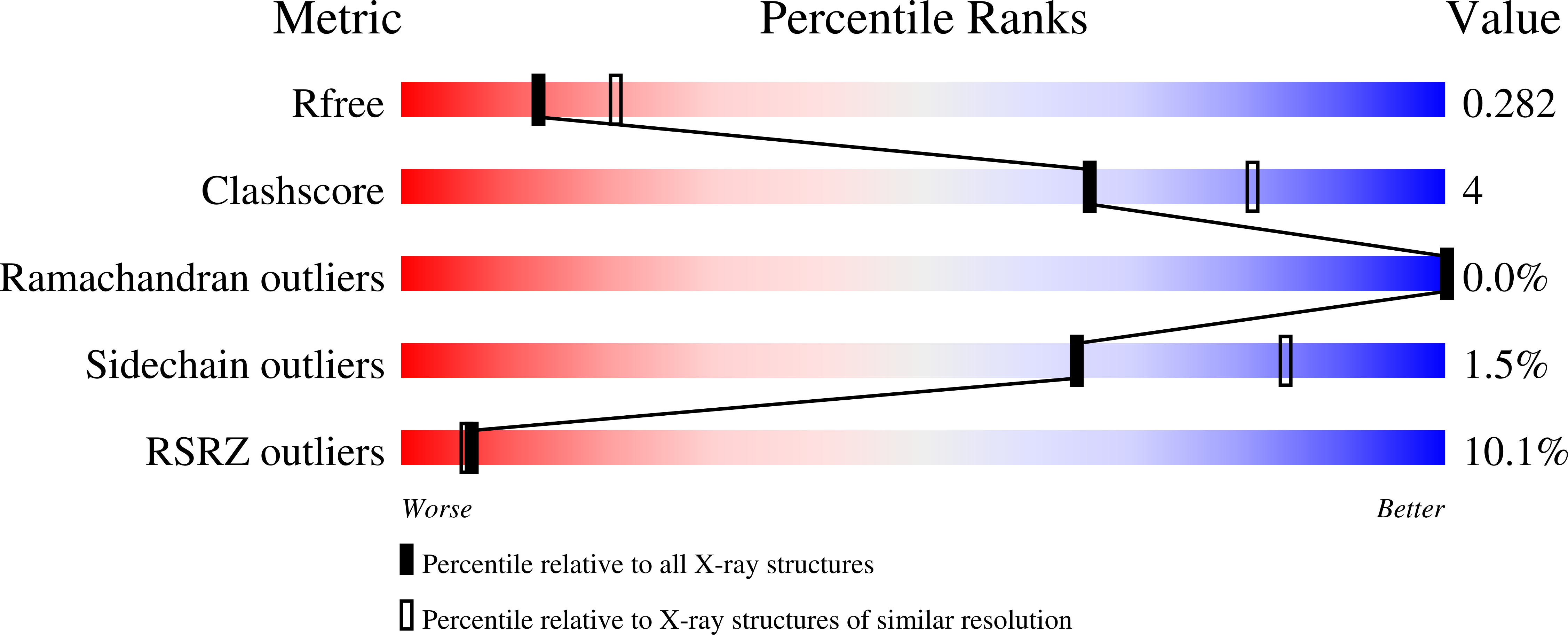
Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
Ye, M., Makola, M., Newman, J.A., Fairhead, M., MacLean, E., Krojer, T., Wright, N.D., Koekemoer, L., Thompson, A., Bezerra, G.A., Yi, G., Li, H., Rangel, V.L., Mamalis, D., Aitkenhead, H., Gilbert, R.J.C., Duerr, K., Davis, B.G., Bountra, C., Gileadi, O., von Delft, F.To be published.