Optimized Ebselen-Based Inhibitors of Bacterial Ureases with Nontypical Mode of Action.
Macegoniuk, K., Tabor, W., Mazzei, L., Cianci, M., Giurg, M., Olech, K., Burda-Grabowska, M., Kaleta, R., Grabowiecka, A., Mucha, A., Ciurli, S., Berlicki, L.(2023) J Med Chem 66: 2054-2063
- PubMed: 36661843
- DOI: https://doi.org/10.1021/acs.jmedchem.2c01799
- Primary Citation of Related Structures:
7ZCY - PubMed Abstract:
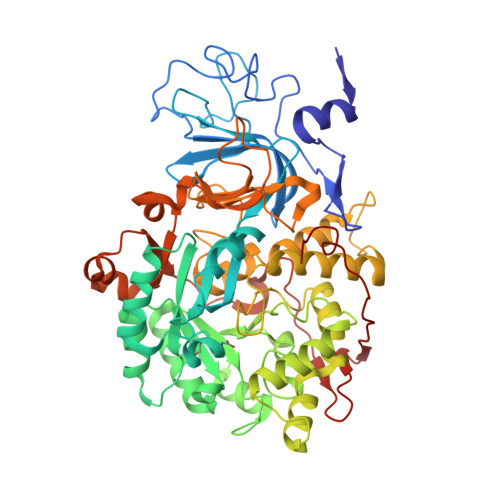
Screening of 25 analogs of Ebselen, diversified at the N-aromatic residue, led to the identification of the most potent inhibitors of Sporosarcina pasteurii urease reported to date. The presence of a dihalogenated phenyl ring caused exceptional activity of these 1,2-benzisoselenazol-3(2 H )-ones, with K i value in a low picomolar range (<20 pM). The affinity was attributed to the increased π-π and π-cation interactions of the dihalogenated phenyl ring with αHis323 and αArg339 during the initial step of binding. Complementary biological studies with selected compounds on the inhibition of ureolysis in whole Proteus mirabilis cells showed a very good potency (IC 50 < 25 nM in phosphate-buffered saline (PBS) buffer and IC 90 < 50 nM in a urine model) for monosubstituted N-phenyl derivatives. The crystal structure of S. pasteurii urease inhibited by one of the most active analogs revealed the recurrent selenation of the Cys322 thiolate, yielding an unprecedented Cys322-S-Se-Se chemical moiety.
Organizational Affiliation:
Department of Bioorganic Chemistry, Wrocław University of Science and Technology, Wybrzeże Wyspiańskiego 27, 50-370 Wrocław, Poland.