Molecular basis of RNA-binding and autoregulation by the cancer-associated splicing factor RBM39.
Campagne, S., Jutzi, D., Malard, F., Matoga, M., Romane, K., Feldmuller, M., Colombo, M., Ruepp, M.D., Allain, F.H.(2023) Nat Commun 14: 5366-5366
- PubMed: 37666821
- DOI: https://doi.org/10.1038/s41467-023-40254-5
- Primary Citation of Related Structures:
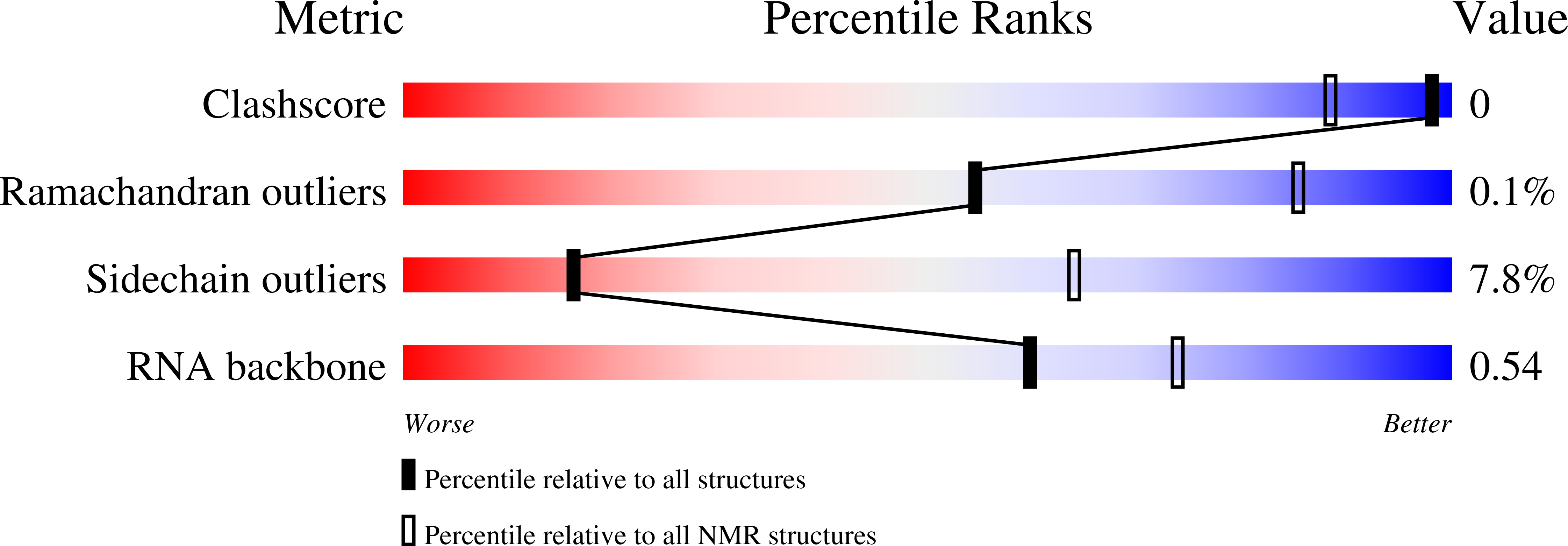
7Q33, 7ZAP - PubMed Abstract:
Pharmacologic depletion of RNA-binding motif 39 (RBM39) using aryl sulfonamides represents a promising anti-cancer therapy but requires high levels of the adaptor protein DCAF15. Consequently, novel approaches to deplete RBM39 in an DCAF15-independent manner are required. Here, we uncover that RBM39 autoregulates via the inclusion of a poison exon into its own pre-mRNA and identify the cis-acting elements that govern this regulation. We also determine the NMR solution structures of RBM39's tandem RNA recognition motifs (RRM1 and RRM2) bound to their respective RNA targets, revealing how RRM1 recognises RNA stem loops whereas RRM2 binds specifically to single-stranded N(G/U)NUUUG. Our results support a model where RRM2 selects the 3'-splice site of a poison exon and the RRM3 and RS domain stabilise the U2 snRNP at the branchpoint. Our work provides molecular insights into RBM39-dependent 3'-splice site selection and constitutes a solid basis to design alternative anti-cancer therapies.
Organizational Affiliation:
ETH Zurich, Department of Biology, Institute of Biochemistry, 8093, Zurich, Switzerland. sebastien.campagne@inserm.fr.