Rational Engineering of an Improved Genetically Encoded pH Sensor Based on Superecliptic pHluorin.
Shen, Y., Wen, Y., Sposini, S., Vishwanath, A.A., Abdelfattah, A.S., Schreiter, E.R., Lemieux, M.J., de Juan-Sanz, J., Perrais, D., Campbell, R.E.(2023) ACS Sens 8: 3014-3022
- PubMed: 37481776
- DOI: https://doi.org/10.1021/acssensors.3c00484
- Primary Citation of Related Structures:
7YV3, 7YV5 - PubMed Abstract:
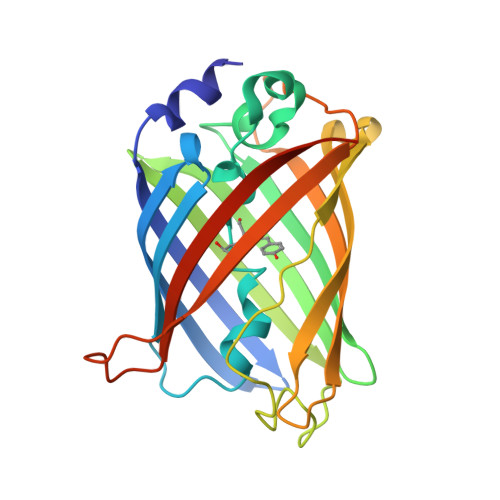
Genetically encoded pH sensors based on fluorescent proteins are valuable tools for the imaging of cellular events that are associated with pH changes, such as exocytosis and endocytosis. Superecliptic pHluorin (SEP) is a pH-sensitive green fluorescent protein (GFP) variant widely used for such applications. Here, we report the rational design, development, structure, and applications of Lime, an improved SEP variant with higher fluorescence brightness and greater pH sensitivity. The X-ray crystal structure of Lime supports the mechanistic rationale that guided the introduction of beneficial mutations. Lime provides substantial improvements relative to SEP for imaging of endocytosis and exocytosis. Furthermore, Lime and its variants are advantageous for a broader range of applications including the detection of synaptic release and neuronal voltage changes.
Organizational Affiliation:
Department of Chemistry, University of Alberta, Edmonton, Alberta T6G 2G2, Canada.