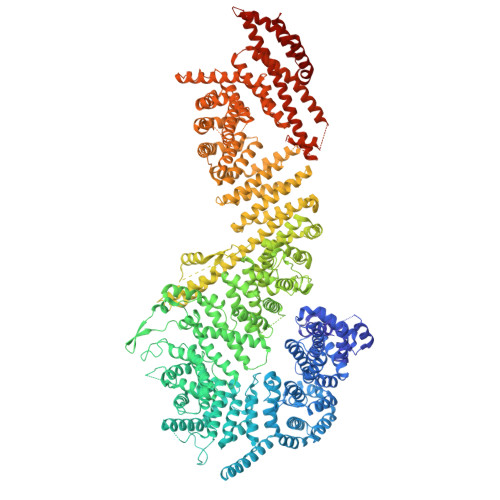
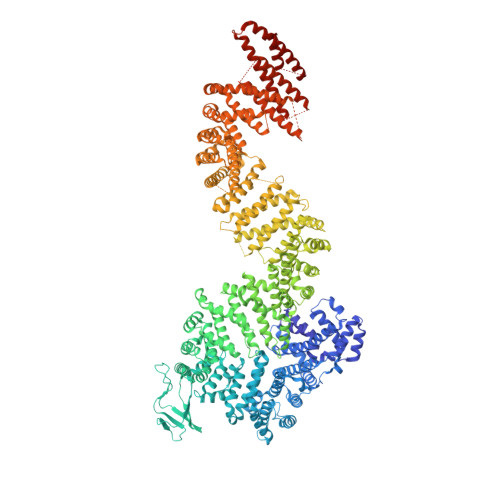
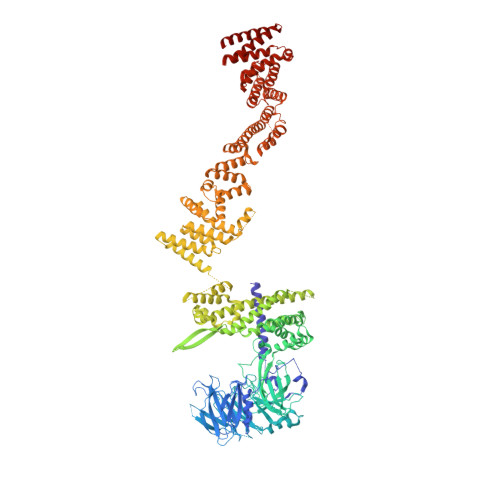
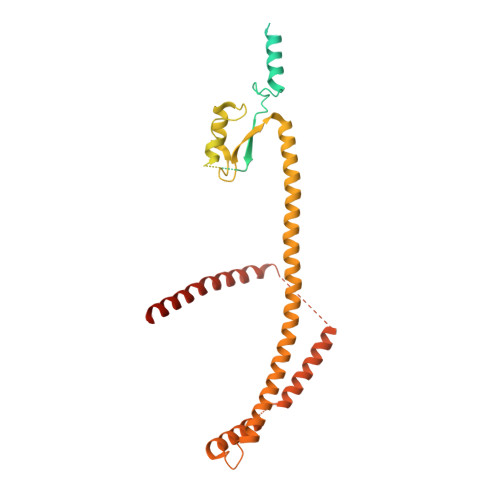
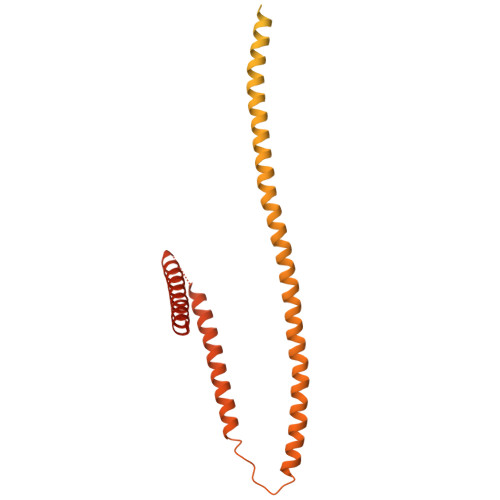
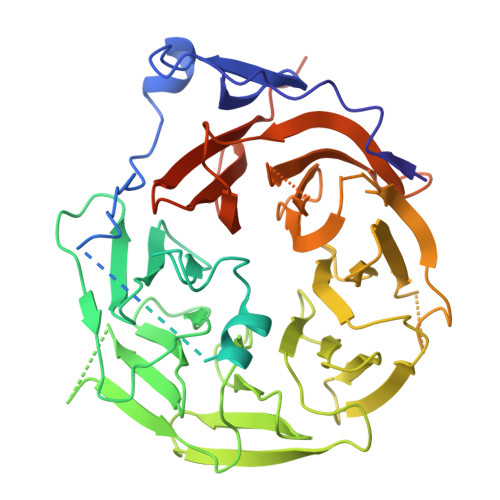
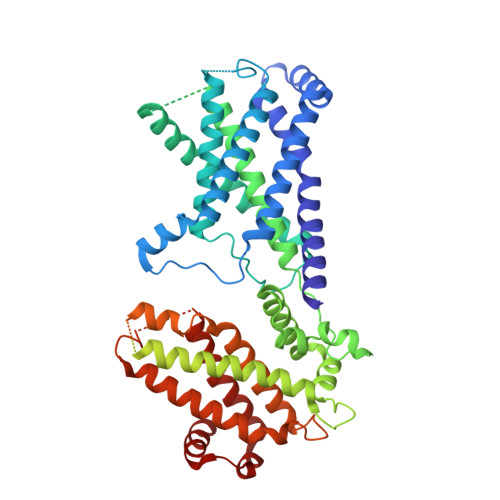
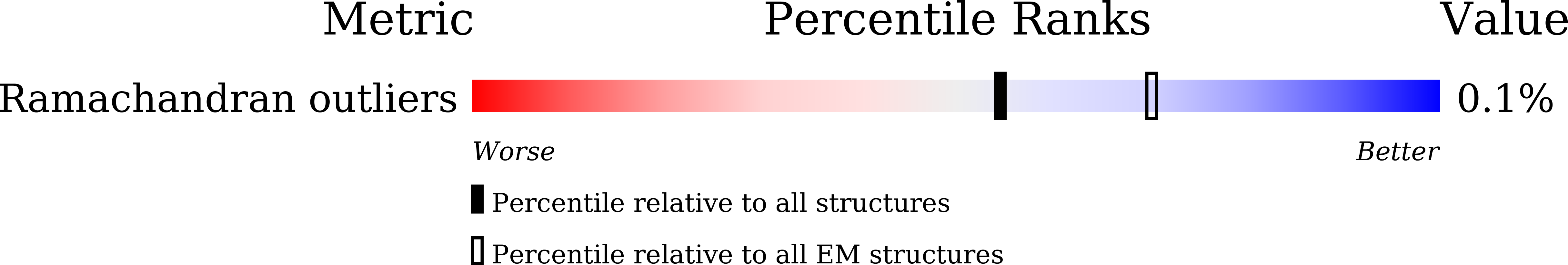Cryo-EM structure of the inner ring from the Xenopus laevis nuclear pore complex.
Huang, G., Zhan, X., Zeng, C., Liang, K., Zhu, X., Zhao, Y., Wang, P., Wang, Q., Zhou, Q., Tao, Q., Liu, M., Lei, J., Yan, C., Shi, Y.(2022) Cell Res 32: 451-460
- PubMed: 35301439
- DOI: https://doi.org/10.1038/s41422-022-00633-x
- Primary Citation of Related Structures:
7WKK - PubMed Abstract:
Nuclear pore complex (NPC) mediates nucleocytoplasmic shuttling. Here we present single-particle cryo-electron microscopy structure of the inner ring (IR) subunit from the Xenopus laevis NPC at an average resolution of 4.2 Å. A homo-dimer of Nup205 resides at the center of the IR subunit, flanked by two molecules of Nup188. Four molecules of Nup93 each places an extended helix into the axial groove of Nup205 or Nup188, together constituting the central scaffold. The channel nucleoporin hetero-trimer of Nup62/58/54 is anchored on the central scaffold. Six Nup155 molecules interact with the central scaffold and together with the NDC1-ALADIN hetero-dimers anchor the IR subunit to the nuclear envelope and to outer rings. The scarce inter-subunit contacts may allow sufficient latitude in conformation and diameter of the IR. Our structure reveals the molecular basis for the IR subunit assembly of a vertebrate NPC.
Organizational Affiliation:
Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang, China. huanggaoxingyu@westlake.edu.cn.