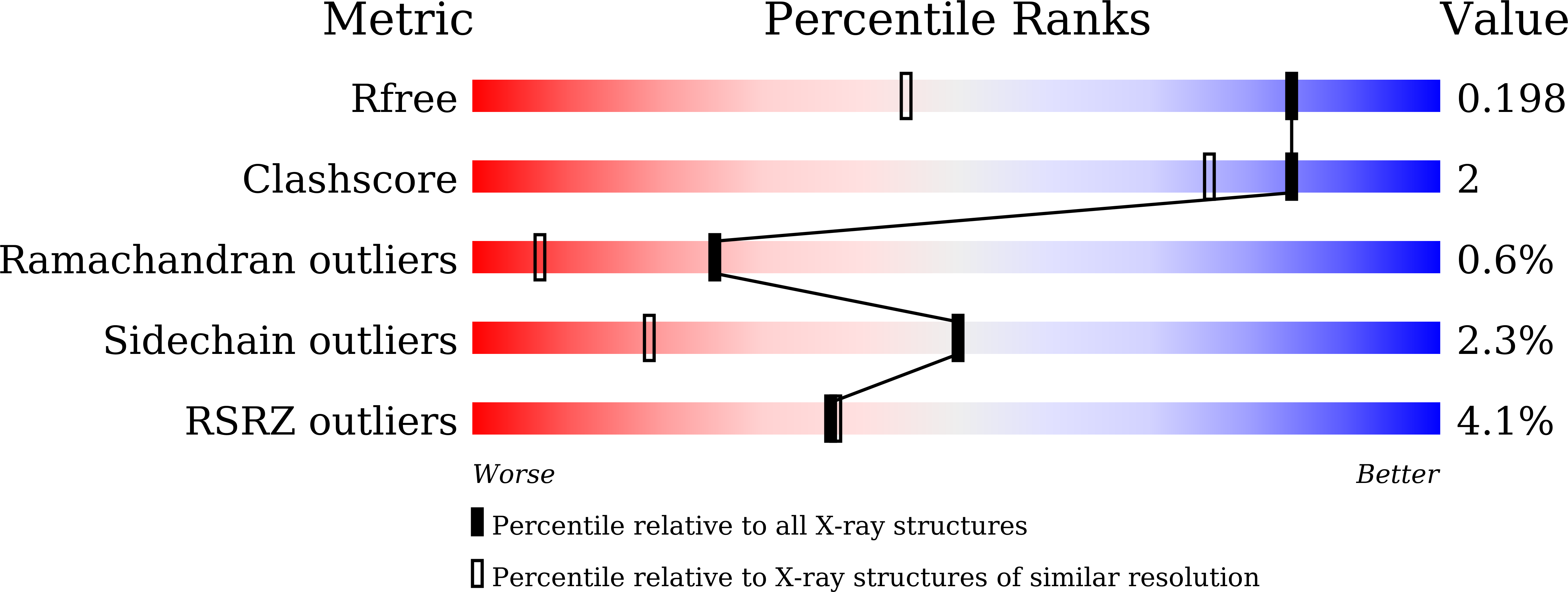
Crystal structure and biochemical analysis of the specialized deoxynivalenol-detoxifying glyoxalase SPG from Gossypium hirsutum.
Hu, Y., Li, H., Min, J., Yu, Y., Liu, W., Huang, J.W., Zhang, L., Yang, Y., Dai, L., Chen, C.C., Guo, R.T.(2022) Int J Biol Macromol 200: 388-396
- PubMed: 35051496
- DOI: https://doi.org/10.1016/j.ijbiomac.2022.01.055
- Primary Citation of Related Structures:
7VQ6 - PubMed Abstract:
Deoxynivalenol (DON) and its acetylated derivatives such as 3-acetyldeoxynivalenol (3A-DON) and 15-acetyldeoxynivalenol (15A-DON) are notorious mycotoxins in Fusarium contaminated cereals, which pose a great threat to human and livestock health. The specialized glyoxalase I from Gossypium hirsutum (SPG) can lower the toxicity of 3A-DON by conducting isomerization to transfer C8 carbonyl to C7 and double bond from C9-C10 to C8-C9. Here we report that the substrate-flexible SPG can also recognize 15A-DON and DON, probably following the same isomerization mechanism as that for 3A-DON. The crystallographic, mutagenesis, and biochemical analyses revealed that SPG provides a hydrophobic pocket to accommodate the substrate and residue E167 might serve as the catalytic base. A variant SPG Y62A that was constructed based on structure-based protein engineering exhibited elevated catalytic activity towards DON, 3A-DON, and 15A-DON by >70%. Furthermore, variant SPG Y62A was successfully expressed in Pichia pastoris, whose catalytic activity was also compared to that produced in Escherichia coli. These results provide a blueprint for further protein engineering of SPG and reveal the potential applications of the enzyme in detoxifying DON, 3A-DON and 15A-DON.
Organizational Affiliation:
State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-Resources, Hubei Key Laboratory of Industrial Biotechnology, Hubei Key Laboratory of Regional Development and Environmental Response, Faculty of Resources and Environmental Science, School of Life Sciences, Hubei University, Wuhan 430062, PR China.