Inhibitory to non-inhibitory evolution of the zeta subunit of the F 1 F O -ATPase of Paracoccus denitrificans and alpha-proteobacteria as related to mitochondrial endosymbiosis.
Mendoza-Hoffmann, F., Yang, L., Buratto, D., Brito-Sanchez, J., Garduno-Javier, G., Salinas-Lopez, E., Uribe-Alvarez, C., Ortega, R., Sotelo-Serrano, O., Cevallos, M.A., Ramirez-Silva, L., Uribe-Carvajal, S., Perez-Hernandez, G., Celis-Sandoval, H., Garcia-Trejo, J.J.(2023) Front Mol Biosci 10: 1184200-1184200
- PubMed: 37664184
- DOI: https://doi.org/10.3389/fmolb.2023.1184200
- Primary Citation of Related Structures:
7VKV - PubMed Abstract:
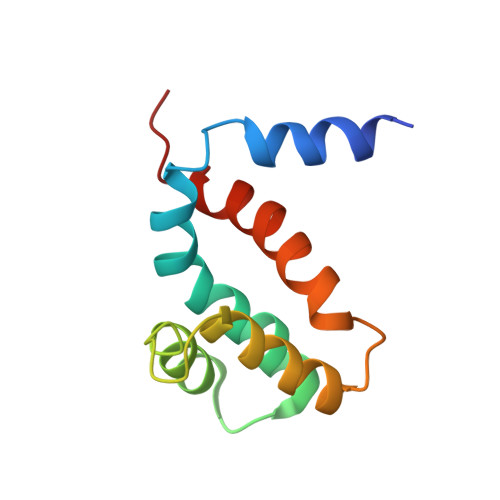
Introduction: The ζ subunit is a potent inhibitor of the F 1 F O -ATPase of Paracoccus denitrificans (PdF 1 F O -ATPase) and related α -proteobacteria different from the other two canonical inhibitors of bacterial ( ε ) and mitochondrial (IF 1 ) F 1 F O -ATPases. ζ mimics mitochondrial IF 1 in its inhibitory N-terminus, blocking the PdF 1 F O -ATPase activity as a unidirectional pawl-ratchet and allowing the PdF 1 F O -ATP synthase turnover. ζ is essential for the respiratory growth of P. denitrificans , as we showed by a Δζ knockout. Given the vital role of ζ in the physiology of P. denitrificans , here, we assessed the evolution of ζ across the α -proteobacteria class. Methods: Through bioinformatic, biochemical, molecular biology, functional, and structural analyses of several ζ subunits, we confirmed the conservation of the inhibitory N-terminus of ζ and its divergence toward its C-terminus. We reconstituted homologously or heterologously the recombinant ζ subunits from several α -proteobacteria into the respective F-ATPases, including free-living photosynthetic, facultative symbiont, and intracellular facultative or obligate parasitic α-proteobacteria. Results and discussion: The results show that ζ evolved, preserving its inhibitory function in free-living α-proteobacteria exposed to broad environmental changes that could compromise the cellular ATP pools. However, the ζ inhibitory function was diminished or lost in some symbiotic α-proteobacteria where ζ is non-essential given the possible exchange of nutrients and ATP from hosts. Accordingly, the ζ gene is absent in some strictly parasitic pathogenic Rickettsiales, which may obtain ATP from the parasitized hosts. We also resolved the NMR structure of the ζ subunit of Sinorhizobium meliloti (Sm- ζ ) and compared it with its structure modeled in AlphaFold. We found a transition from a compact ordered non-inhibitory conformation into an extended α-helical inhibitory N-terminus conformation, thus explaining why the Sm- ζ cannot exert homologous inhibition. However, it is still able to inhibit the PdF 1 F O -ATPase heterologously. Together with the loss of the inhibitory function of α-proteobacterial ε , the data confirm that the primary inhibitory function of the α-proteobacterial F 1 F O -ATPase was transferred from ε to ζ and that ζ, ε, and IF 1 evolved by convergent evolution. Some key evolutionary implications on the endosymbiotic origin of mitochondria, as most likely derived from α -proteobacteria, are also discussed.
Organizational Affiliation:
Departamento de Biología, Facultad de Química, Ciudad Universitaria, Universidad Nacional Autónoma de México (U.N.A.M.), Ciudad de México, México.