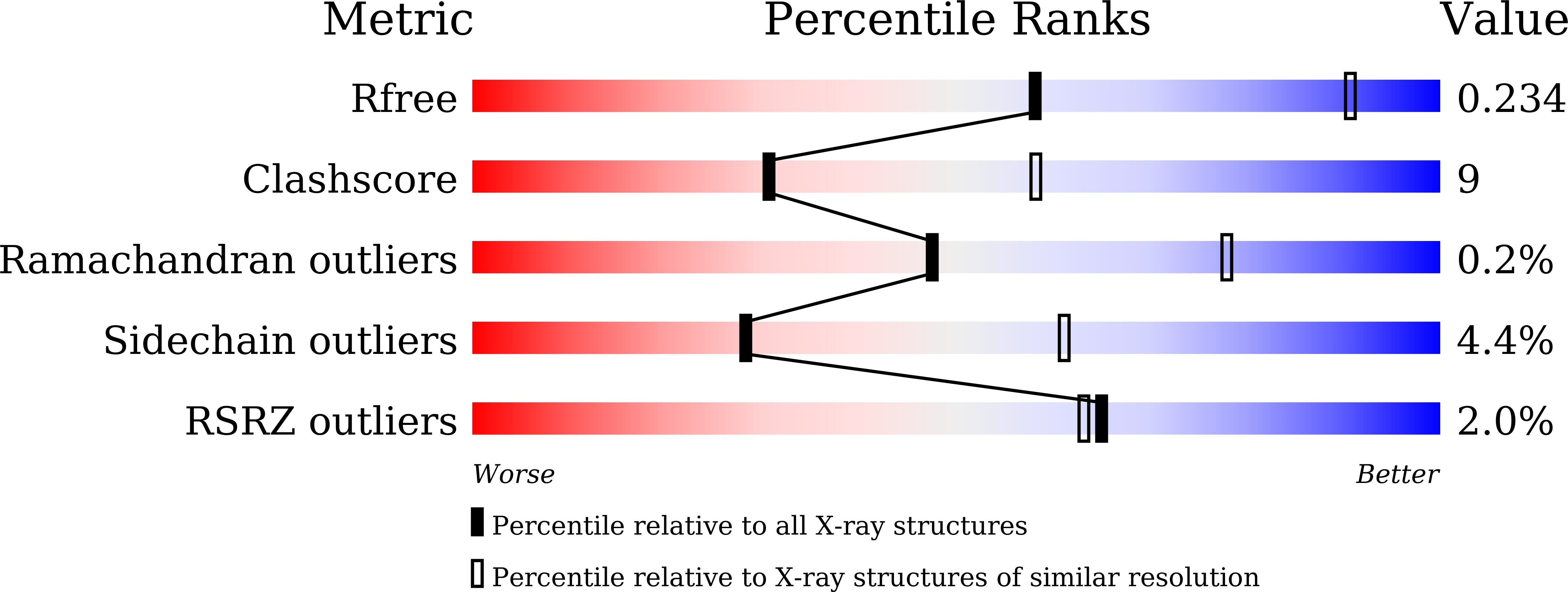
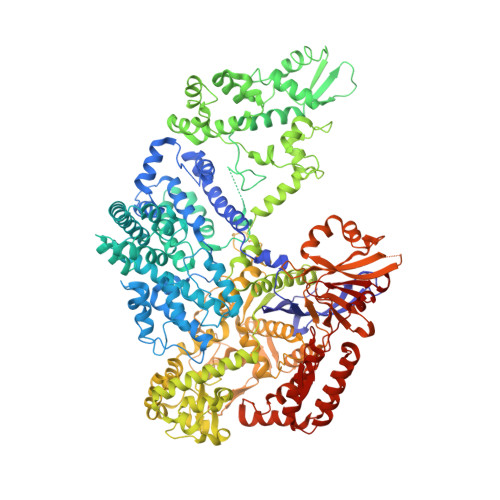
Structural and Dynamics Studies of the Spcas9 Variant Provide Insights into the Regulatory Role of the REC1 Domain
Liu, H., Zhou, Y., Song, Y., Zhang, Q., Kan, Y., Tang, X., Xiao, Q., Xiang, Q., Liu, H., Luo, Y., Bao, R.(2022) ACS Catal 12: 8687-8697