Crystallographic characterization of a marine invertebrate ferritin from the sea cucumber Apostichopus japonicus.
Wu, Y., Ming, T., Huo, C., Qiu, X., Su, C., Lu, C., Zhou, J., Li, Y., Su, X.(2022) FEBS Open Bio 12: 664-674
- PubMed: 35090095
- DOI: https://doi.org/10.1002/2211-5463.13375
- Primary Citation of Related Structures:
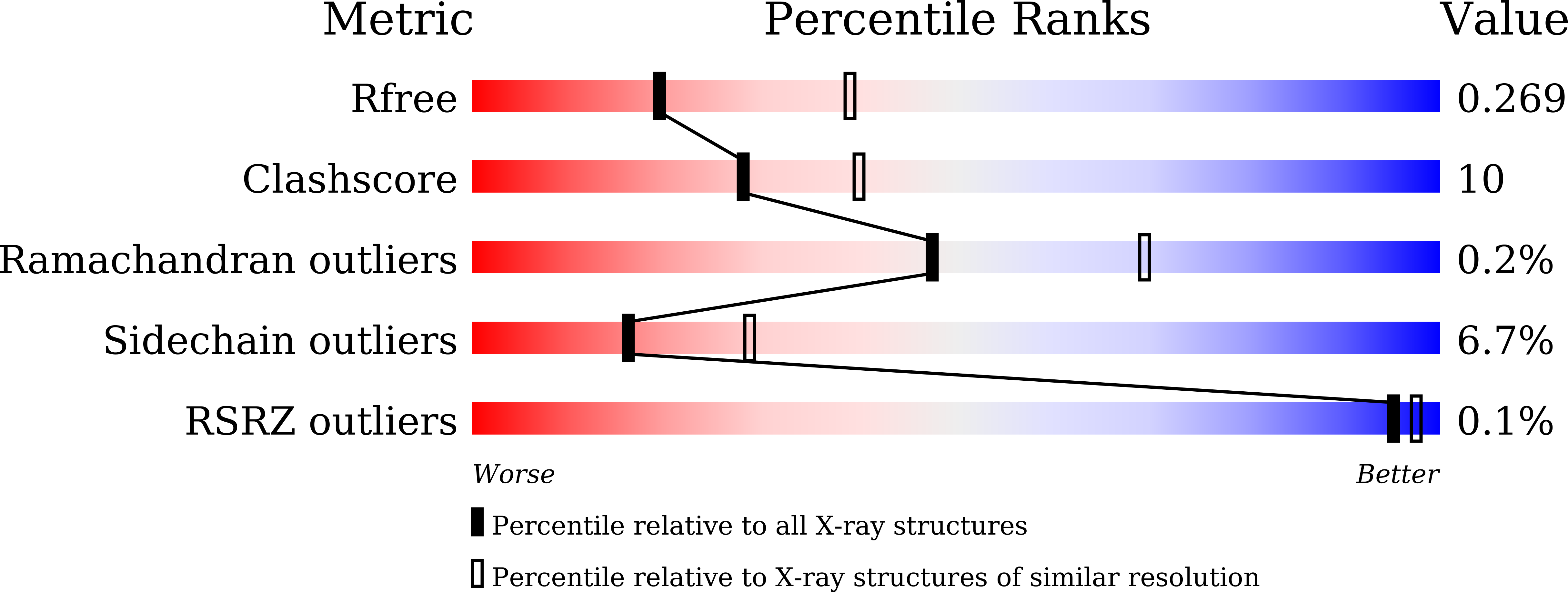
7VHR - PubMed Abstract:
Ferritin is considered to be an ubiquitous and conserved iron-binding protein that plays a crucial role in iron storage, detoxification, and immune response. Although ferritin is of critical importance for almost all kingdoms of life, there is a lack of knowledge about its role in the marine invertebrate sea cucumber (Apostichopus japonicus). In this study, we characterized the first crystal structure of A. japonicus ferritin (AjFER) at 2.75 Å resolution. The structure of AjFER shows a 4-3-2 symmetry cage-like hollow shell composed of 24 subunits, mostly similar to the structural characteristics of other known ferritin species, including the conserved ferroxidase center and 3-fold channel. The 3-fold channel consisting of three 3-fold negative amino acid rings suggests a potential pathway in which metal ions can be first captured by Asp120 from the outside environment, attracted by His116 and Cys128 when entering the channel, and then transferred by Glu138 from the 3-fold channel to the ferroxidase site. Overall, the presented crystal structure of AjFER may provide insights into the potential mechanism of the metal transport pathway for related marine invertebrate ferritins.
Organizational Affiliation:
State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Ningbo University, China.