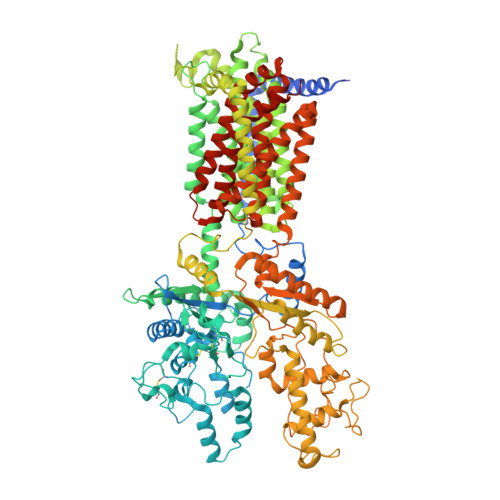
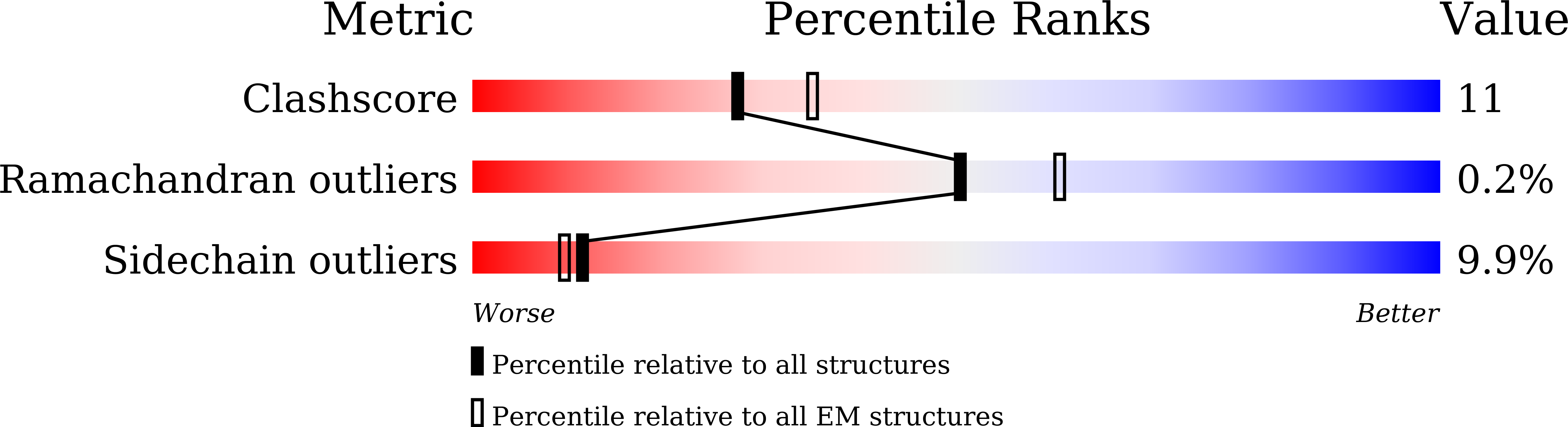Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Luo, Y., Wan, G., Zhang, X., Zhou, X., Wang, Q., Fan, J., Cai, H., Ma, L., Wu, H., Qu, Q., Cong, Y., Zhao, Y., Li, D.(2021) Structure 29: 1286
- PubMed: 34174188
- DOI: https://doi.org/10.1016/j.str.2021.06.004
- Primary Citation of Related Structures:
7V6Y, 7V6Z - PubMed Abstract:
The 12-transmembrane protein Patched (Ptc1) acts as a suppressor for Hedgehog (Hh) signaling by depleting sterols in the cytoplasmic membrane leaflet that are required for the activation of downstream regulators. The positive modulator Hh inhibits Ptc1's transporter function by binding to Ptc1 and its co-receptors, which are locally concentrated in invaginated microdomains known as caveolae. Here, we reconstitute the mouse Ptc1 into lipid nanodiscs and determine its structure using single-particle cryoelectron microscopy. The structure is overall similar to those in amphipol and detergents but displays various conformational differences in the transmembrane region. Although most particles show monomers, we observe Ptc1 dimers with distinct interaction patterns and different membrane curvatures, some of which are reminiscent of caveolae. We find that an extramembranous "hand-shake" region rich in hydrophobic and aromatic residues mediates inter-Ptc1 interactions under different membrane curvatures. Our data provide a plausible framework for Ptc1 clustering in the highly curved caveolae.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, State Key Laboratory of Cell Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China; School of Life Science and Technology, ShanghaiTech University, 393 Middle Huaxia Road, Shanghai 201210, China.