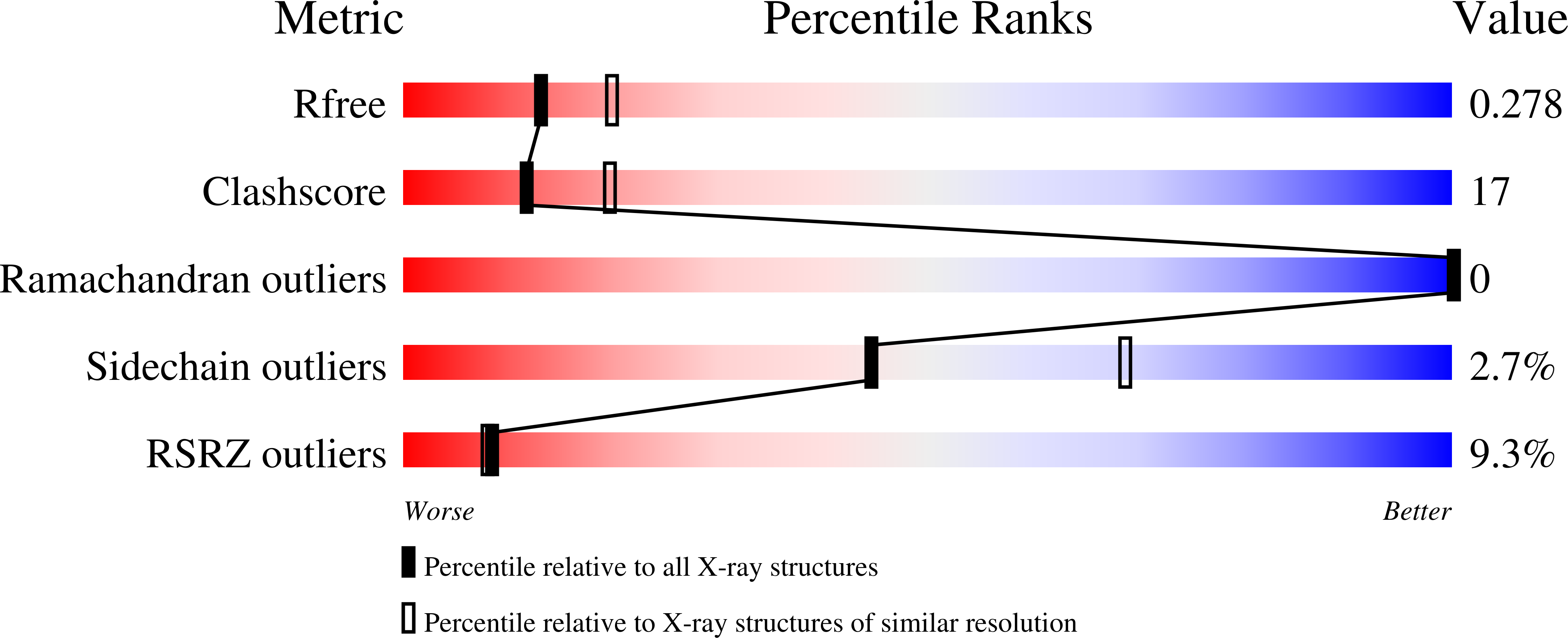
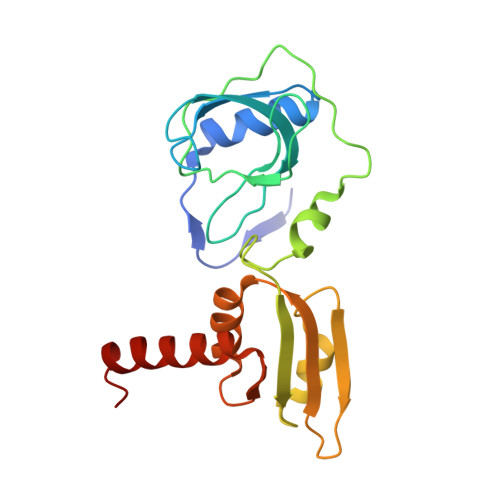
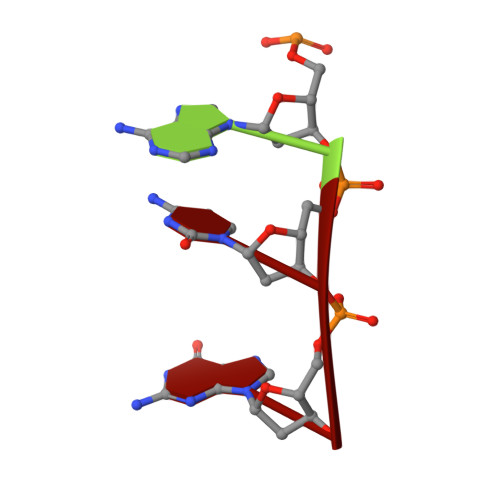
ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Gozzi, K., Salinas, R., Nguyen, V.D., Laub, M.T., Schumacher, M.A.(2022) Genes Dev 36: 618-633
- PubMed: 35618312
- DOI: https://doi.org/10.1101/gad.349541.122
- Primary Citation of Related Structures:
7TZV, 7U02 - PubMed Abstract:
DNA damage repair systems are critical for genomic integrity. However, they must be coordinated with DNA replication and cell division to ensure accurate genomic transmission. In most bacteria, this coordination is mediated by the SOS response through LexA, which triggers a halt in cell division until repair is completed. Recently, an SOS-independent damage response system was revealed in Caulobacter crescentus. This pathway is controlled by the transcription activator, DriD, but how DriD senses and signals DNA damage is unknown. To address this question, we performed biochemical, cellular, and structural studies. We show that DriD binds a specific promoter DNA site via its N-terminal HTH domain to activate transcription of genes, including the cell division inhibitor didA A structure of the C-terminal portion of DriD revealed a WYL motif domain linked to a WCX dimerization domain. Strikingly, we found that DriD binds ssDNA between the WYL and WCX domains. Comparison of apo and ssDNA-bound DriD structures reveals that ssDNA binding orders and orients the DriD domains, indicating a mechanism for ssDNA-mediated operator DNA binding activation. Biochemical and in vivo studies support the structural model. Our data thus reveal the molecular mechanism underpinning an SOS-independent DNA damage repair pathway.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology. Cambridge, Massachusetts 02139, USA.