Crystal structure of R14A human Galectin-7 mutant in presence of 4-O-beta-D-Galactopyranosyl-D-glucose
Pham, N.T.H., Calmettes, C., Doucet, N.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Galectin-7 | 135 | Homo sapiens | Mutation(s): 1 Gene Names: LGALS7, PIG1, LGALS7B | 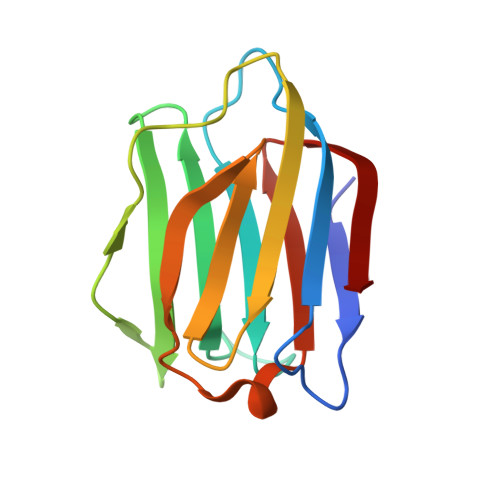 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P47929 (Homo sapiens) Explore P47929 Go to UniProtKB: P47929 | |||||
PHAROS: P47929 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P47929 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LBL (Subject of Investigation/LOI) Query on LBL | C [auth A], G [auth B] | (2~{R},3~{R},4~{R},5~{R})-4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2,3,5,
6-tetrakis(oxidanyl)hexanal C12 H22 O11 DKXNBNKWCZZMJT-JVCRWLNRSA-N |  | ||
| GOL Query on GOL | F [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | D [auth A], E [auth A], H [auth B], I [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 30.25 | α = 90 |
| b = 76.38 | β = 90 |
| c = 111.3 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Natural Sciences and Engineering Research Council (NSERC, Canada) | RGPIN 2016-05557 | |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | R01GM105978 | |
| Other government | Fonds de Recherche du Quebec - Sante (FRQ-S) - Research Scholar Senior Career Award (281993) | |
| Other government | Fonds de Recherche du Quebec - Sante (FRQ-S) - Junior 1 (251848) | |
| Natural Sciences and Engineering Research Council (NSERC, Canada) | RGPIN-2017-06091 | |
| Other government | Fonds de Recherche du Quebec - Sante (FRQ-S) - Doctoral Training scholarship (287239) |