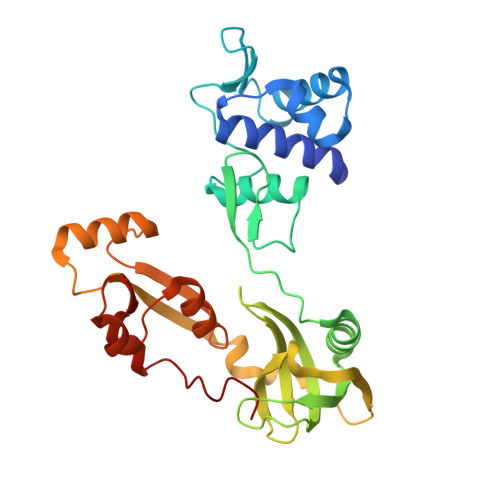
Identification and characterization of the WYL BrxR protein and its gene as separable regulatory elements of a BREX phage restriction system.
Luyten, Y.A., Hausman, D.E., Young, J.C., Doyle, L.A., Higashi, K.M., Ubilla-Rodriguez, N.C., Lambert, A.R., Arroyo, C.S., Forsberg, K.J., Morgan, R.D., Stoddard, B.L., Kaiser, B.K.(2022) Nucleic Acids Res 50: 5171-5190
- PubMed: 35511079
- DOI: https://doi.org/10.1093/nar/gkac311
- Primary Citation of Related Structures:
7T8K, 7T8L - PubMed Abstract:
Bacteriophage exclusion ('BREX') phage restriction systems are found in a wide range of bacteria. Various BREX systems encode unique combinations of proteins that usually include a site-specific methyltransferase; none appear to contain a nuclease. Here we describe the identification and characterization of a Type I BREX system from Acinetobacter and the effect of deleting each BREX ORF on growth, methylation, and restriction. We identified a previously uncharacterized gene in the BREX operon that is dispensable for methylation but involved in restriction. Biochemical and crystallographic analyses of this factor, which we term BrxR ('BREX Regulator'), demonstrate that it forms a homodimer and specifically binds a DNA target site upstream of its transcription start site. Deletion of the BrxR gene causes cell toxicity, reduces restriction, and significantly increases the expression of BrxC. In contrast, the introduction of a premature stop codon into the BrxR gene, or a point mutation blocking its DNA binding ability, has little effect on restriction, implying that the BrxR coding sequence and BrxR protein play independent functional roles. We speculate that elements within the BrxR coding sequence are involved in cis regulation of anti-phage activity, while the BrxR protein itself plays an additional regulatory role, perhaps during horizontal transfer.
Organizational Affiliation:
New England Biolabs, 240 County Road, Ipswich, MA 01938, USA.