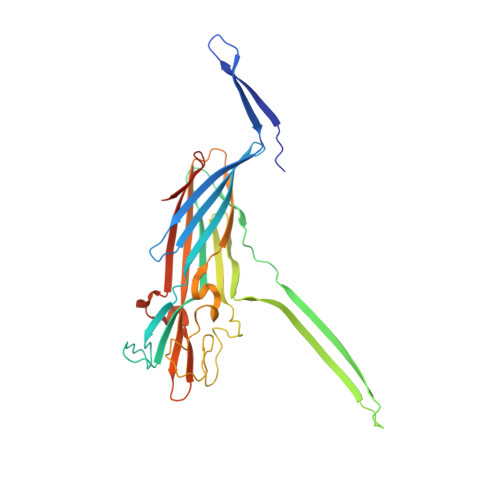
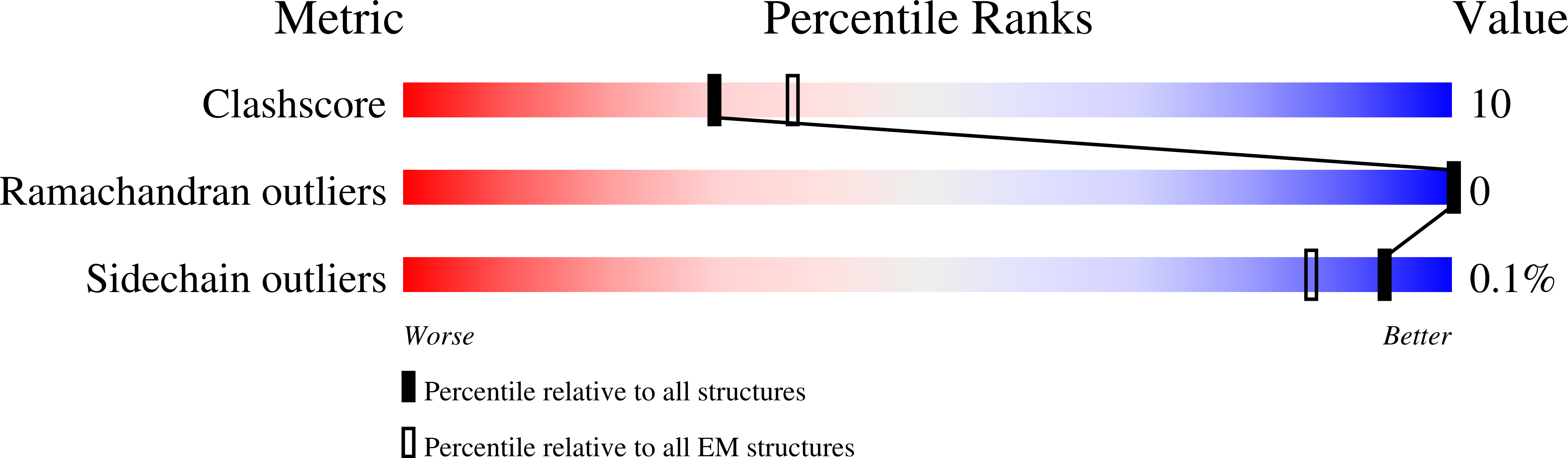Emerging enterococcus pore-forming toxins with MHC/HLA-I as receptors.
Xiong, X., Tian, S., Yang, P., Lebreton, F., Bao, H., Sheng, K., Yin, L., Chen, P., Zhang, J., Qi, W., Ruan, J., Wu, H., Chen, H., Breault, D.T., Wu, H., Earl, A.M., Gilmore, M.S., Abraham, J., Dong, M.(2022) Cell 185: 1157
- PubMed: 35259335
- DOI: https://doi.org/10.1016/j.cell.2022.02.002
- Primary Citation of Related Structures:
7T4D, 7T4E - PubMed Abstract:
Enterococci are a part of human microbiota and a leading cause of multidrug resistant infections. Here, we identify a family of Enterococcus pore-forming toxins (Epxs) in E. faecalis, E. faecium, and E. hirae strains isolated across the globe. Structural studies reveal that Epxs form a branch of β-barrel pore-forming toxins with a β-barrel protrusion (designated the top domain) sitting atop the cap domain. Through a genome-wide CRISPR-Cas9 screen, we identify human leukocyte antigen class I (HLA-I) complex as a receptor for two members (Epx2 and Epx3), which preferentially recognize human HLA-I and homologous MHC-I of equine, bovine, and porcine, but not murine, origin. Interferon exposure, which stimulates MHC-I expression, sensitizes human cells and intestinal organoids to Epx2 and Epx3 toxicity. Co-culture with Epx2-harboring E. faecium damages human peripheral blood mononuclear cells and intestinal organoids, and this toxicity is neutralized by an Epx2 antibody, demonstrating the toxin-mediated virulence of Epx-carrying Enterococcus.
Organizational Affiliation:
Department of Urology, Boston Children's Hospital, Department of Surgery, Harvard Medical School, Boston, MA 02115, USA; Department of Microbiology, Harvard Medical School, Boston, MA 02115, USA.