Molecular insights into the HLA-B35 molecules' classification associated with HIV control.
Lobos, C.A., Chatzileontiadou, D.S., Sok, B., Almedia, C.A., Halim, H., D'Orsogna, L., Gras, S.(2024) Immunol Cell Biol 102: 34-45
- PubMed: 37811811
- DOI: https://doi.org/10.1111/imcb.12698
- Primary Citation of Related Structures:
7SIF, 7SIG, 7SIH - PubMed Abstract:
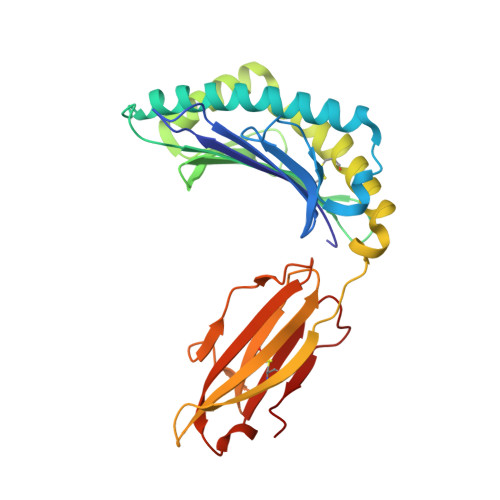
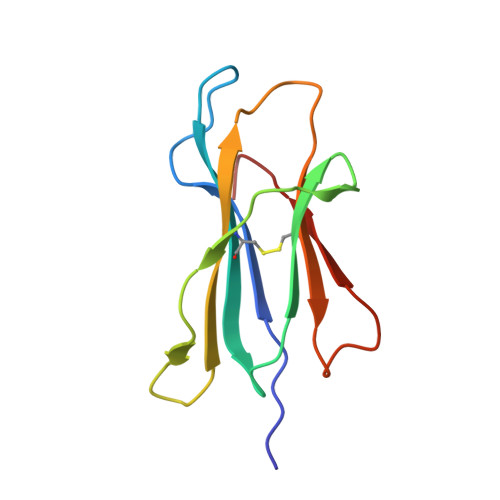
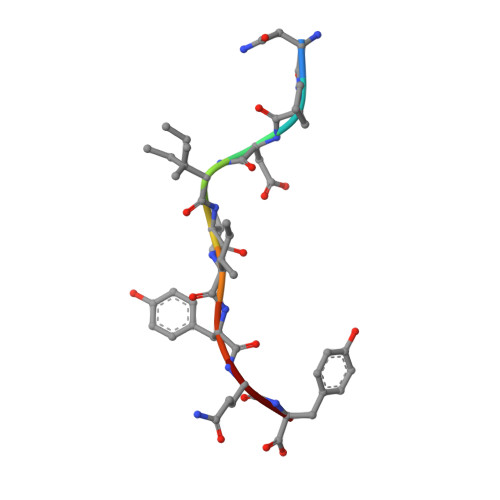
Human leukocyte antigen (HLA) class I molecules have been shown to influence the immune response to HIV infection and acquired immunodeficiency syndrome progression. Polymorphisms within the HLA-B35 molecules divide the family into two groups, namely, Px and PY. The Px group is associated with deleterious effects and accelerated disease progression in HIV + patients, whereas the PY group is not. The classification is based on the preferential binding of a tyrosine at the C-terminal part of the peptide in the PY group, and a nontyrosine residue in the Px group. However, there is a lack of knowledge on the molecular differences between the two groups. Here, we have investigated three HLA-B35 molecules, namely, HLA-B*35:01 (PY), HLA-B*35:03 (Px) and HLA-B*35:05 (unclassified). We selected an HIV-derived peptide, NY9, and demonstrated that it can trigger a polyfunctional CD8 + T-cell response in HLA-B*35:01 + /HIV + patients. We determined that in the complex with the NY9 peptide, the PY molecule was more stable than the Px molecule. We solved the crystal structures of the three HLA molecules in complex with the NY9 peptide, and structural similarities with HLA-B*35:01 would classify the HLA-B*35:05 within the PY group. Interestingly, we found that HLA-B*35:05 can also bind a small molecule in its cleft, suggesting that small drugs could bind as well.
Organizational Affiliation:
Department of Biochemistry and Chemistry, La Trobe Institute for Molecular Science, La Trobe University, Bundoora, VIC, Australia.