Discovery of Inhibitors of the Lipopolysaccharide Transporter MsbA: From a Screening Hit to Potent Wild-Type Gram-Negative Activity.
Verma, V.A., Wang, L., Labadie, S.S., Liang, J., Sellers, B.D., Wang, J., Dong, L., Wang, Q., Zhang, S., Xu, Z., Zhang, Y., Niu, Y., Wang, X., Wai, J., Koehler, M.F.T., Hu, H., Alexander, M.K., Nishiyama, M., Miu, A., Xu, Y., Pang, J., Katakam, A.K., Reichelt, M., Austin, C.D., Ho, H., Payandeh, J., Koth, C.M.(2022) J Med Chem 65: 4085-4120
- PubMed: 35184554
- DOI: https://doi.org/10.1021/acs.jmedchem.1c01909
- Primary Citation of Related Structures:
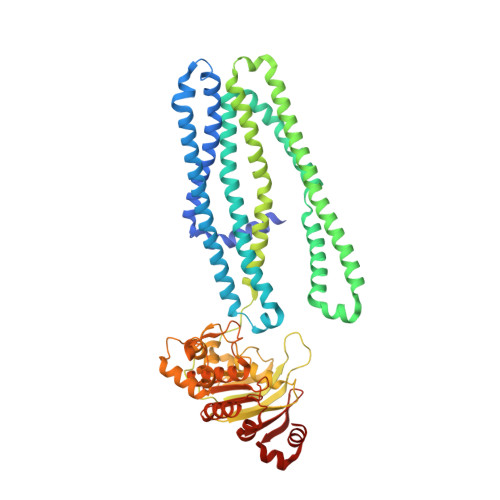
7SEL - PubMed Abstract:
The dramatic increase in the prevalence of multi-drug resistant Gram-negative bacterial infections and the simultaneous lack of new classes of antibiotics is projected to result in approximately 10 million deaths per year by 2050. We report on efforts to target the Gram-negative ATP-binding cassette (ABC) transporter MsbA, an essential inner membrane protein that transports lipopolysaccharide from the inner leaflet to the periplasmic face of the inner membrane. We demonstrate the improvement of a high throughput screening hit into compounds with on-target single digit micromolar (μM) minimum inhibitory concentrations against wild-type uropathogenic Escherichia coli , Klebsiella pneumoniae , and Enterobacter cloacae . A 2.98 Å resolution X-ray crystal structure of MsbA complexed with an inhibitor revealed a novel mechanism for inhibition of an ABC transporter. The identification of a fully encapsulated membrane binding site in Gram-negative bacteria led to unique physicochemical property requirements for wild-type activity.
Organizational Affiliation:
Genentech, Inc., 1 DNA Way, South San Francisco, California 94080, United States.