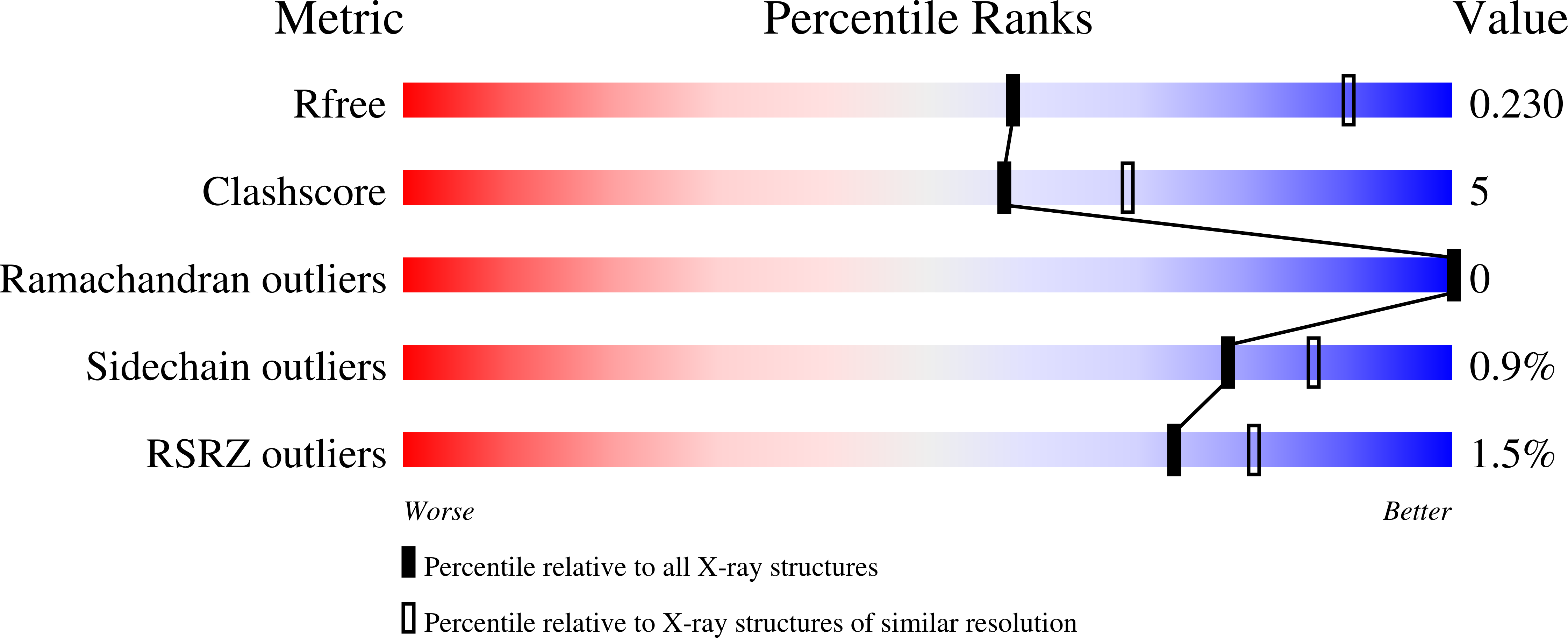
Crystal Structure of Dihydroorotate dehydrogenase from Plasmodium falciparum in complex with Orotate, FMN, and inhibitor NCGC00600348-01
Seattle Structural Genomics Center for Infectious Disease (SSGCID), Shek, R., Abendroth, J., Lorimer, D., Edwards, T.E., Eastman, R., Van Voorhis, W.To be published.