Serial macromolecular crystallography at ALBA Synchrotron Light Source.
Martin-Garcia, J.M., Botha, S., Hu, H., Jernigan, R., Castellvi, A., Lisova, S., Gil, F., Calisto, B., Crespo, I., Roy-Chowdhury, S., Grieco, A., Ketawala, G., Weierstall, U., Spence, J., Fromme, P., Zatsepin, N., Boer, D.R., Carpena, X.(2022) J Synchrotron Radiat 29: 896-907
- PubMed: 35511023
- DOI: https://doi.org/10.1107/S1600577522002508
- Primary Citation of Related Structures:
7S4R, 7S4W, 7S4Y, 7S4Z, 7S50 - PubMed Abstract:
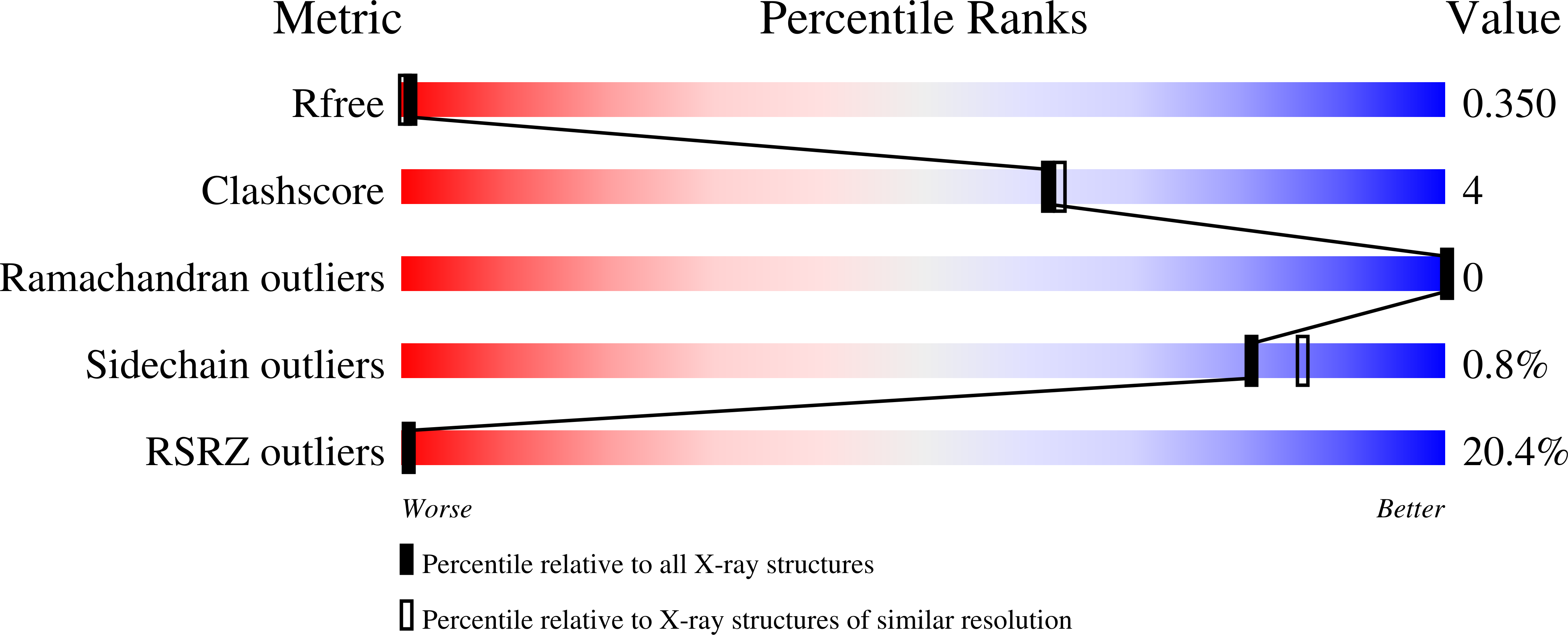
The increase in successful adaptations of serial crystallography at synchrotron radiation sources continues. To date, the number of serial synchrotron crystallography (SSX) experiments has grown exponentially, with over 40 experiments reported so far. In this work, we report the first SSX experiments with viscous jets conducted at ALBA beamline BL13-XALOC. Small crystals (15-30 µm) of five soluble proteins (lysozyme, proteinase K, phycocyanin, insulin and α-spectrin-SH3 domain) were suspended in lipidic cubic phase (LCP) and delivered to the X-ray beam with a high-viscosity injector developed at Arizona State University. Complete data sets were collected from all proteins and their high-resolution structures determined. The high quality of the diffraction data collected from all five samples, and the lack of specific radiation damage in the structures obtained in this study, confirm that the current capabilities at the beamline enables atomic resolution determination of protein structures from microcrystals as small as 15 µm using viscous jets at room temperature. Thus, BL13-XALOC can provide a feasible alternative to X-ray free-electron lasers when determining snapshots of macromolecular structures.
Organizational Affiliation:
Center for Applied Structural Discovery, Biodesign Institute, Arizona State University, Tempe, AZ, USA.