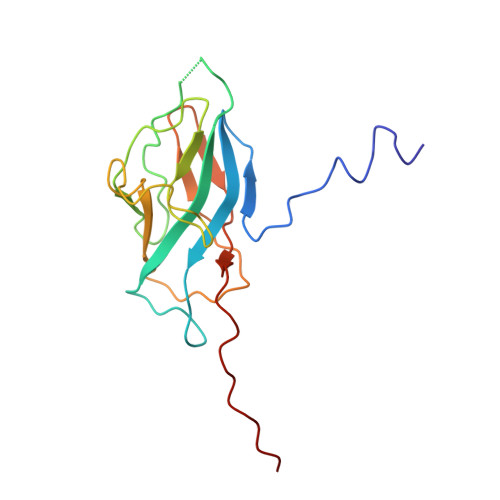
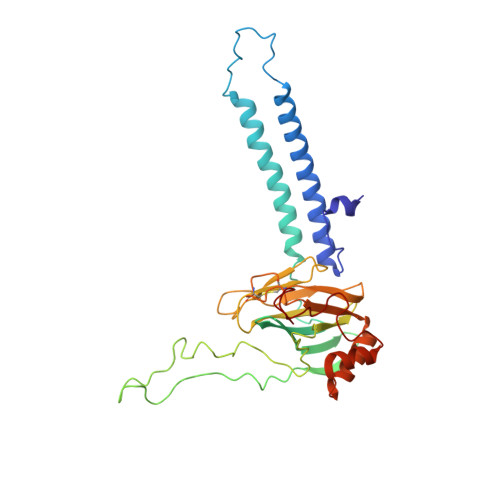
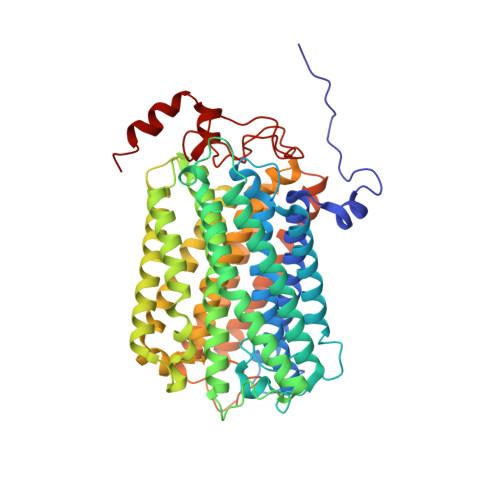
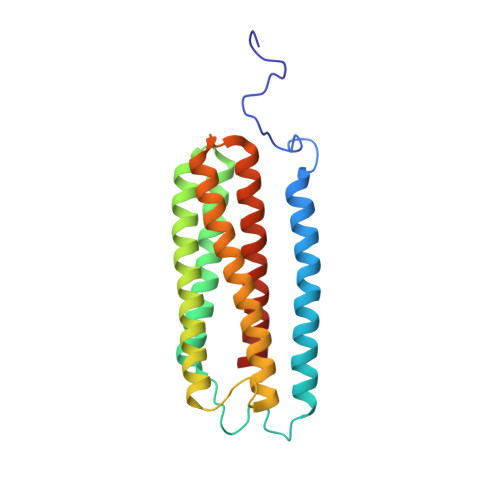
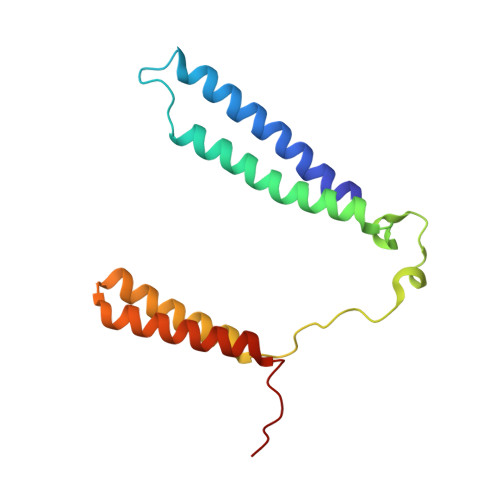
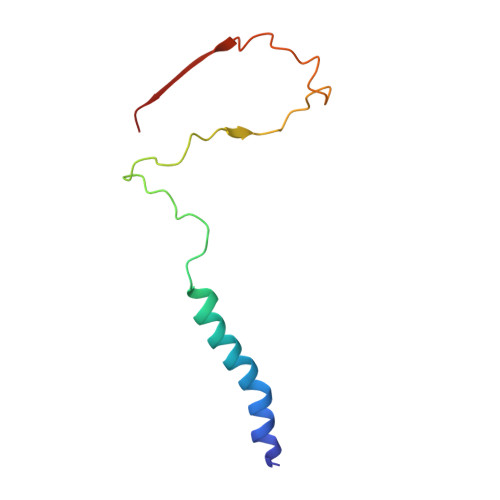
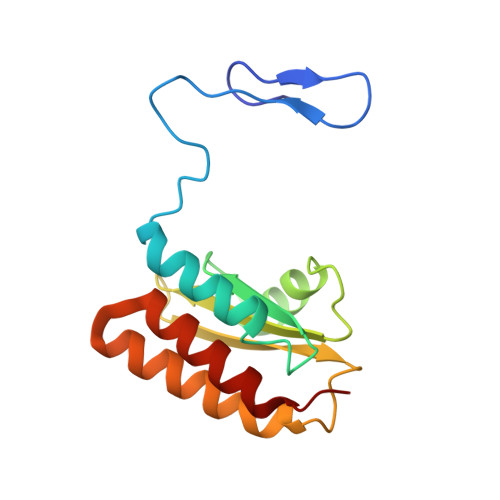
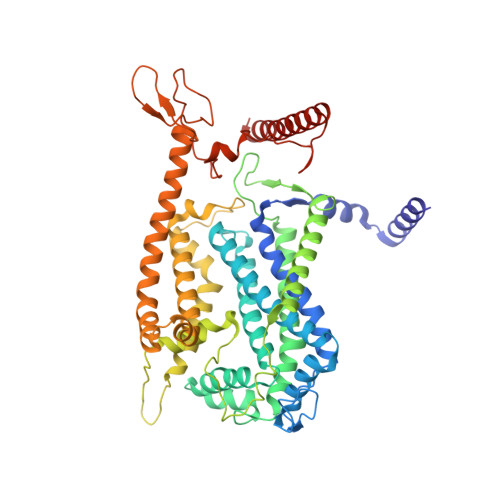
Structure of mycobacterial CIII 2 CIV 2 respiratory supercomplex bound to the tuberculosis drug candidate telacebec (Q203).
Yanofsky, D.J., Di Trani, J.M., Krol, S., Abdelaziz, R., Bueler, S.A., Imming, P., Brzezinski, P., Rubinstein, J.L.(2021) Elife 10
- PubMed: 34590581
- DOI: https://doi.org/10.7554/eLife.71959
- Primary Citation of Related Structures:
7RH5, 7RH6, 7RH7 - PubMed Abstract:
The imidazopyridine telacebec, also known as Q203, is one of only a few new classes of compounds in more than 50 years with demonstrated antituberculosis activity in humans. Telacebec inhibits the mycobacterial respiratory supercomplex composed of complexes III and IV (CIII 2 CIV 2 ). In mycobacterial electron transport chains, CIII 2 CIV 2 replaces canonical CIII and CIV, transferring electrons from the intermediate carrier menaquinol to the final acceptor, molecular oxygen, while simultaneously transferring protons across the inner membrane to power ATP synthesis. We show that telacebec inhibits the menaquinol:oxygen oxidoreductase activity of purified Mycobacterium smegmatis CIII 2 CIV 2 at concentrations similar to those needed to inhibit electron transfer in mycobacterial membranes and Mycobacterium tuberculosis growth in culture. We then used electron cryomicroscopy (cryoEM) to determine structures of CIII 2 CIV 2 both in the presence and absence of telacebec. The structures suggest that telacebec prevents menaquinol oxidation by blocking two different menaquinol binding modes to prevent CIII 2 CIV 2 activity.
Organizational Affiliation:
Molecular Medicine Program, The Hospital for Sick Children, Toronto, Canada.