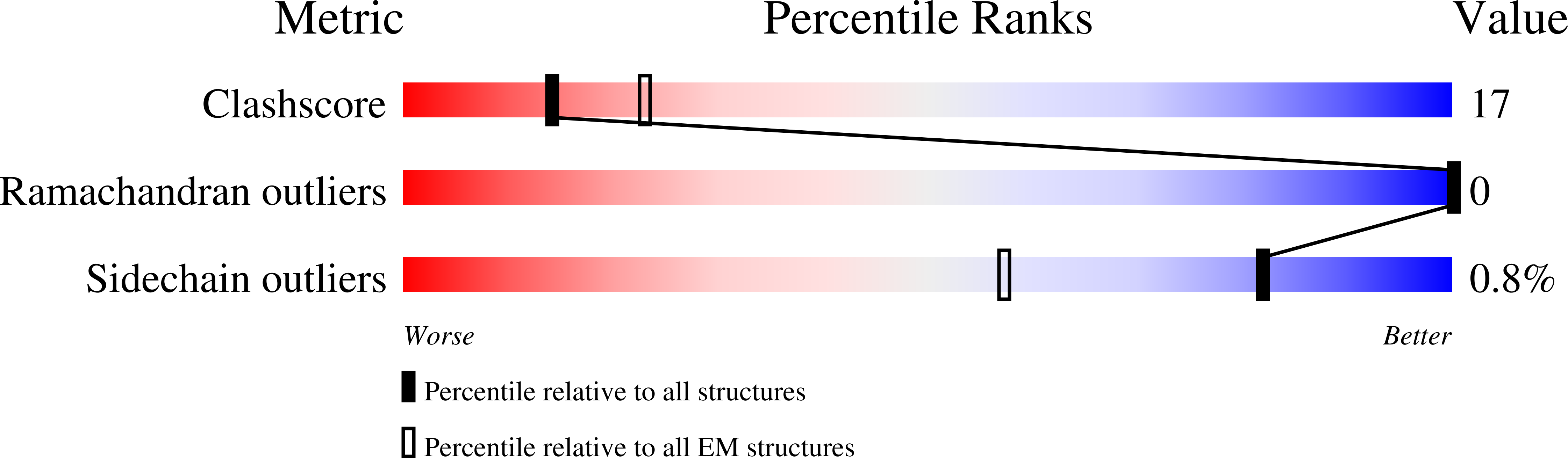Structural determinants of dual incretin receptor agonism by tirzepatide.
Sun, B., Willard, F.S., Feng, D., Alsina-Fernandez, J., Chen, Q., Vieth, M., Ho, J.D., Showalter, A.D., Stutsman, C., Ding, L., Suter, T.M., Dunbar, J.D., Carpenter, J.W., Mohammed, F.A., Aihara, E., Brown, R.A., Bueno, A.B., Emmerson, P.J., Moyers, J.S., Kobilka, T.S., Coghlan, M.P., Kobilka, B.K., Sloop, K.W.(2022) Proc Natl Acad Sci U S A 119: e2116506119-e2116506119
- PubMed: 35333651
- DOI: https://doi.org/10.1073/pnas.2116506119
- Primary Citation of Related Structures:
7RA3, 7RBT, 7RG9, 7RGP - PubMed Abstract:
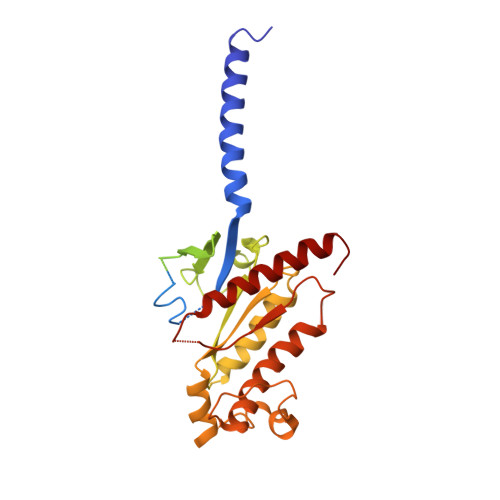
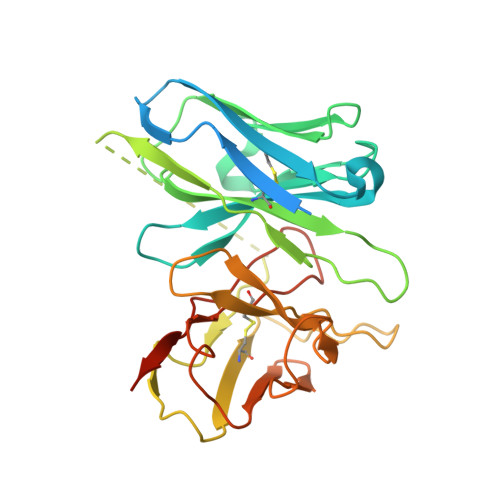
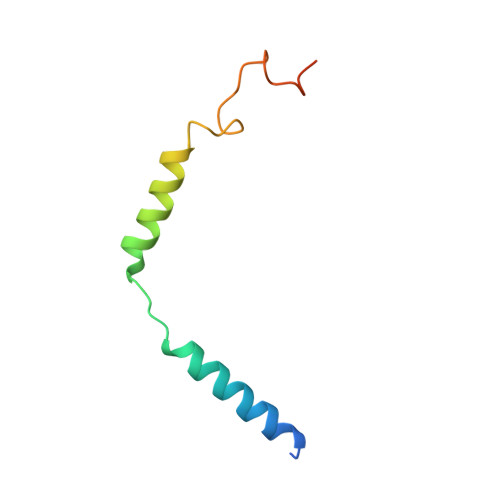
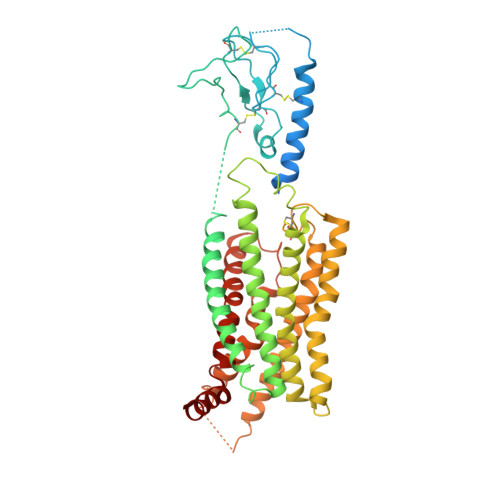
SignificanceTirzepatide is a dual agonist of the glucose-dependent insulinotropic polypeptide receptor (GIPR) and the glucagon-like peptide-1 receptor (GLP-1R), which are incretin receptors that regulate carbohydrate metabolism. This investigational agent has proven superior to selective GLP-1R agonists in clinical trials in subjects with type 2 diabetes mellitus. Intriguingly, although tirzepatide closely resembles native GIP in how it activates the GIPR, it differs markedly from GLP-1 in its activation of the GLP-1R, resulting in less agonist-induced receptor desensitization. We report how cryogenic electron microscopy and molecular dynamics simulations inform the structural basis for the unique pharmacology of tirzepatide. These studies reveal the extent to which fatty acid modification, combined with amino acid sequence, determines the mode of action of a multireceptor agonist.
Organizational Affiliation:
ConfometRx, Santa Clara, CA 95054.