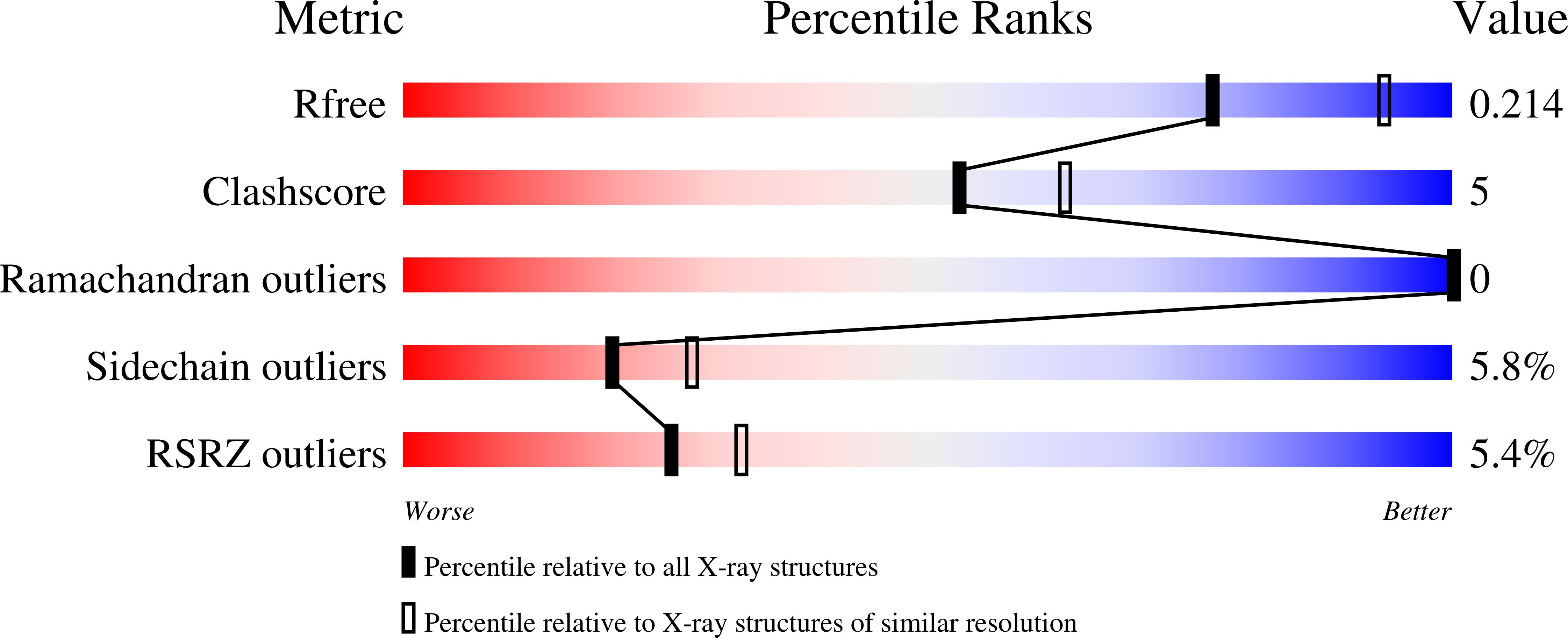
Structural variations between small alarmone hydrolase dimers support different modes of regulation of the stringent response.
Bisiak, F., Chrenkova, A., Zhang, S.D., Pedersen, J.N., Otzen, D.E., Zhang, Y.E., Brodersen, D.E.(2022) J Biol Chem 298: 102142-102142
- PubMed: 35714769
- DOI: https://doi.org/10.1016/j.jbc.2022.102142
- Primary Citation of Related Structures:
7QOC, 7QOD, 7QOE - PubMed Abstract:
The bacterial stringent response involves wide-ranging metabolic reprogramming aimed at increasing long-term survivability during stress conditions. One of the hallmarks of the stringent response is the production of a set of modified nucleotides, known as alarmones, which affect a multitude of cellular pathways in diverse ways. Production and degradation of these molecules depend on the activity of enzymes from the RelA/SpoT homologous family, which come in both bifunctional (containing domains to both synthesize and hydrolyze alarmones) and monofunctional (consisting of only synthetase or hydrolase domain) variants, of which the structure, activity, and regulation of the bifunctional RelA/SpoT homologs have been studied most intensely. Despite playing an important role in guanosine nucleotide homeostasis in particular, mechanisms of regulation of the small alarmone hydrolases (SAHs) are still rather unclear. Here, we present crystal structures of SAH enzymes from Corynebacterium glutamicum (RelH Cg ) and Leptospira levettii (RelH Ll ) and show that while being highly similar, structural differences in substrate access and dimer conformations might be important for regulating their activity. We propose that a varied dimer form is a general property of the SAH family, based on current structural information as well as prediction models for this class of enzymes. Finally, subtle structural variations between monofunctional and bifunctional enzymes point to how these different classes of enzymes are regulated.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Aarhus University, Aarhus C, Denmark.