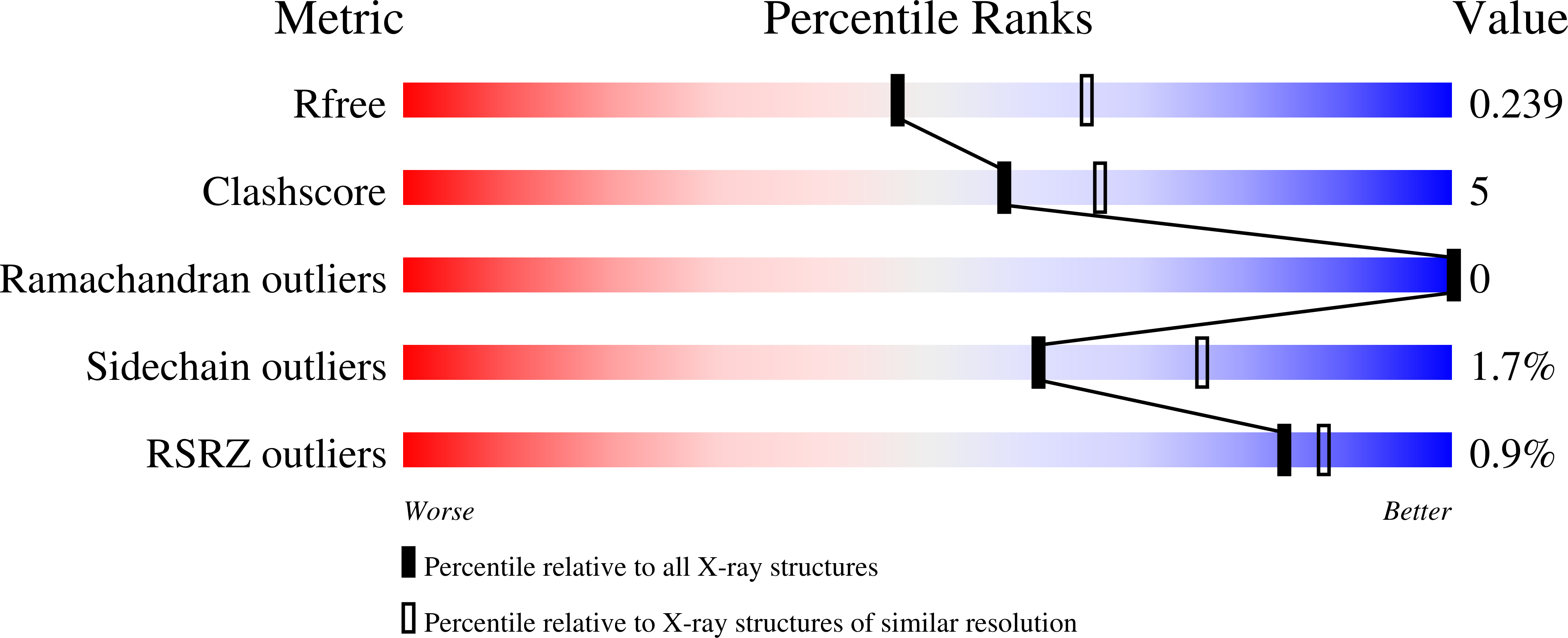
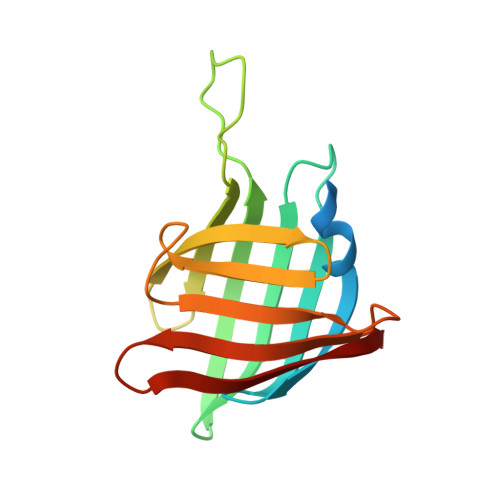
Crystal structures of Streptomyces tsukubaensis sigma factor SigG1 and anti-sigma RsfG.
Leite, J.P., Lourenco, F., Oliveira, R., Sousa, S.F., Mendes, M.V., Gales, L.(2023) J Struct Biol 215: 108038-108038
- PubMed: 37858875
- DOI: https://doi.org/10.1016/j.jsb.2023.108038
- Primary Citation of Related Structures:
7QH3, 7QH5 - PubMed Abstract:
Transcription of specific genes in bacteria under environmental stress is frequently initiated by extracytoplasmic function (ECF) σ factors. ECFs σ factors harbour two conserved domains, σ 2 and σ 4 , for transcription initiation by recognition of the promoter region and recruitment of RNA polymerase (RNAP). The crystal structure of Streptomyces tsukubaensis SigG1, an ECF56-family σ factor, was determined revealing σ 2 , σ 4 and the additional carboxi-terminal domain SnoaL_2 tightly packed in a compact conformation. The structure of anti-sigma RsfG was also determined by X-ray crystallography and shows a rare β-barrel fold. Analysis of the metal binding motifs inside the protein barrel are consistent with Fe(III) binding, which is in agreement with previous findings that the Streptomyces tsukubaensis ECF56 SigG1-RsfG system is involved in metal-ion homeostasis.
Organizational Affiliation:
i3S - Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Rua Alfredo Allen 208, Porto, Porto 4200-135, Portugal; IBMC - Instituto de Biologia Molecular e Celular, Universidade do Porto, Rua Alfredo Allen 208, Porto, Porto 4200-135, Portugal; ICBAS - Instituto de Ciências Biomédicas Abel Salazar, Universidade do Porto, Rua Jorge de Viterbo Ferreira 228, Porto, Porto 4050-313, Portugal.