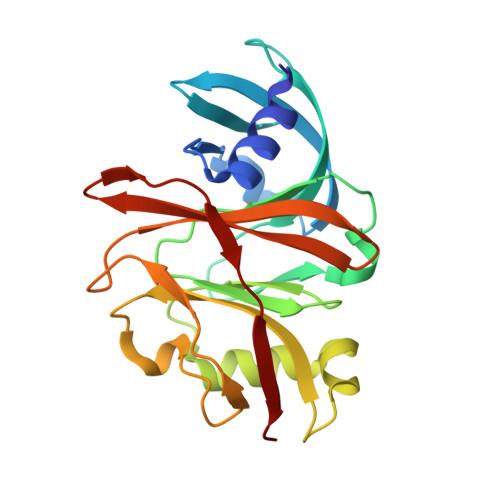
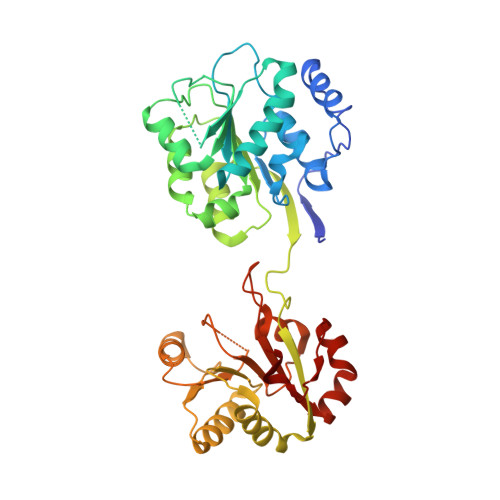
Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Mobbs, G.W., Aziz, A.A., Dix, S.R., Blackburn, G.M., Sedelnikova, S.E., Minshull, T.C., Dickman, M.J., Baker, P.J., Nathan, S., Raih, M.F., Rice, D.W.(2022) Commun Biol 5: 272-272
- PubMed: 35347220
- DOI: https://doi.org/10.1038/s42003-022-03186-2
- Primary Citation of Related Structures:
6RVU, 7PPZ, 7PQ0 - PubMed Abstract:
Burkholderia pseudomallei lethal factor 1 (BLF1) exhibits site-specific glutamine deamidase activity against the eukaryotic RNA helicase, eIF4A, thereby blocking mammalian protein synthesis. The structure of a complex between BLF1 C94S and human eIF4A shows that the toxin binds in the cleft between the two RecA-like eIF4A domains forming interactions with residues from both and with the scissile amide of the target glutamine, Gln339, adjacent to the toxin active site. The RecA-like domains adopt a radically twisted orientation compared to other eIF4A structures and the nature and position of conserved residues suggests this may represent a conformation associated with RNA binding. Comparison of the catalytic site of BLF1 with other deamidases and cysteine proteases reveals that they fall into two classes, related by pseudosymmetry, that present either the re or si faces of the target amide/peptide to the nucleophilic sulfur, highlighting constraints in the convergent evolution of their Cys-His active sites.
Organizational Affiliation:
Krebs Institute, School of Biosciences, University of Sheffield, Sheffield, S10 2TN, UK.