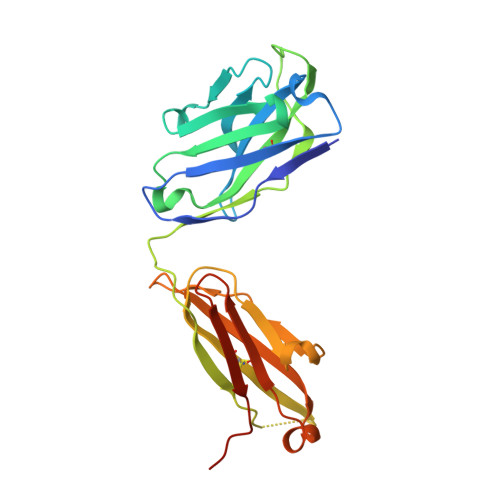
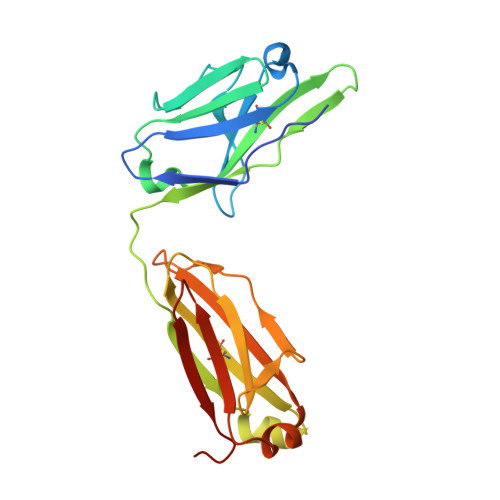
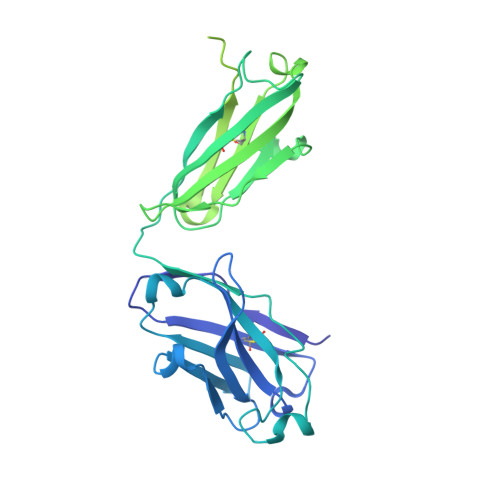
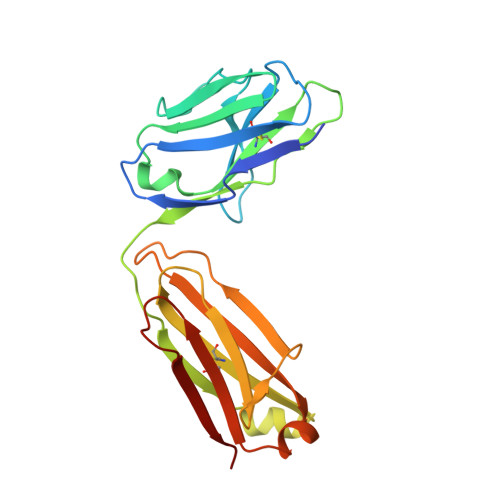
Heterotypic interactions drive antibody synergy against a malaria vaccine candidate.
Ragotte, R.J., Pulido, D., Lias, A.M., Quinkert, D., Alanine, D.G.W., Jamwal, A., Davies, H., Nacer, A., Lowe, E.D., Grime, G.W., Illingworth, J.J., Donat, R.F., Garman, E.F., Bowyer, P.W., Higgins, M.K., Draper, S.J.(2022) Nat Commun 13: 933-933
- PubMed: 35177602
- DOI: https://doi.org/10.1038/s41467-022-28601-4
- Primary Citation of Related Structures:
7PHU, 7PHV, 7PHW, 7PI2, 7PI3, 7PI7 - PubMed Abstract:
Understanding mechanisms of antibody synergy is important for vaccine design and antibody cocktail development. Examples of synergy between antibodies are well-documented, but the mechanisms underlying these relationships often remain poorly understood. The leading blood-stage malaria vaccine candidate, CyRPA, is essential for invasion of Plasmodium falciparum into human erythrocytes. Here we present a panel of anti-CyRPA monoclonal antibodies that strongly inhibit parasite growth in in vitro assays. Structural studies show that growth-inhibitory antibodies bind epitopes on a single face of CyRPA. We also show that pairs of non-competing inhibitory antibodies have strongly synergistic growth-inhibitory activity. These antibodies bind to neighbouring epitopes on CyRPA and form lateral, heterotypic interactions which slow antibody dissociation. We predict that such heterotypic interactions will be a feature of many immune responses. Immunogens which elicit such synergistic antibody mixtures could increase the potency of vaccine-elicited responses to provide robust and long-lived immunity against challenging disease targets.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, Oxford, UK.