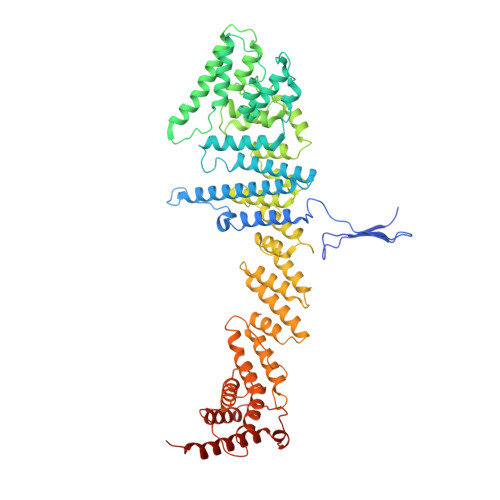
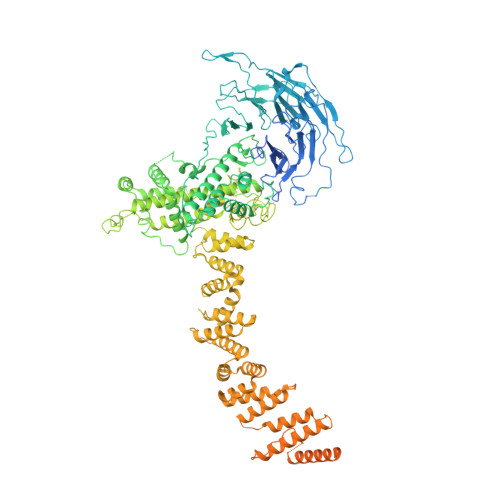
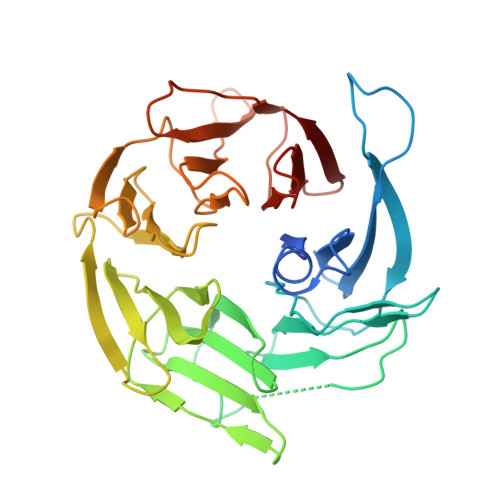
The cellular environment shapes the nuclear pore complex architecture.
Schuller, A.P., Wojtynek, M., Mankus, D., Tatli, M., Kronenberg-Tenga, R., Regmi, S.G., Dip, P.V., Lytton-Jean, A.K.R., Brignole, E.J., Dasso, M., Weis, K., Medalia, O., Schwartz, T.U.(2021) Nature 598: 667-671
- PubMed: 34646014
- DOI: https://doi.org/10.1038/s41586-021-03985-3
- Primary Citation of Related Structures:
7PEQ, 7PER - PubMed Abstract:
Nuclear pore complexes (NPCs) create large conduits for cargo transport between the nucleus and cytoplasm across the nuclear envelope (NE) 1-3 . These multi-megadalton structures are composed of about thirty different nucleoporins that are distributed in three main substructures (the inner, cytoplasmic and nucleoplasmic rings) around the central transport channel 4-6 . Here we use cryo-electron tomography on DLD-1 cells that were prepared using cryo-focused-ion-beam milling to generate a structural model for the human NPC in its native environment. We show that-compared with previous human NPC models obtained from purified NEs-the inner ring in our model is substantially wider; the volume of the central channel is increased by 75% and the nucleoplasmic and cytoplasmic rings are reorganized. Moreover, the NPC membrane exhibits asymmetry around the inner-ring complex. Using targeted degradation of Nup96, a scaffold nucleoporin of the cytoplasmic and nucleoplasmic rings, we observe the interdependence of each ring in modulating the central channel and maintaining membrane asymmetry. Our findings highlight the inherent flexibility of the NPC and suggest that the cellular environment has a considerable influence on NPC dimensions and architecture.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA.