Discovery of Dimeric Arylsulfonamides as Potent ADAM8 Inhibitors.
Cuffaro, D., Camodeca, C., Tuccinardi, T., Ciccone, L., Bartsch, J.W., Kellermann, T., Cook, L., Nuti, E., Rossello, A.(2021) ACS Med Chem Lett 12: 1787-1793
- PubMed: 35111280
- DOI: https://doi.org/10.1021/acsmedchemlett.1c00411
- Primary Citation of Related Structures:
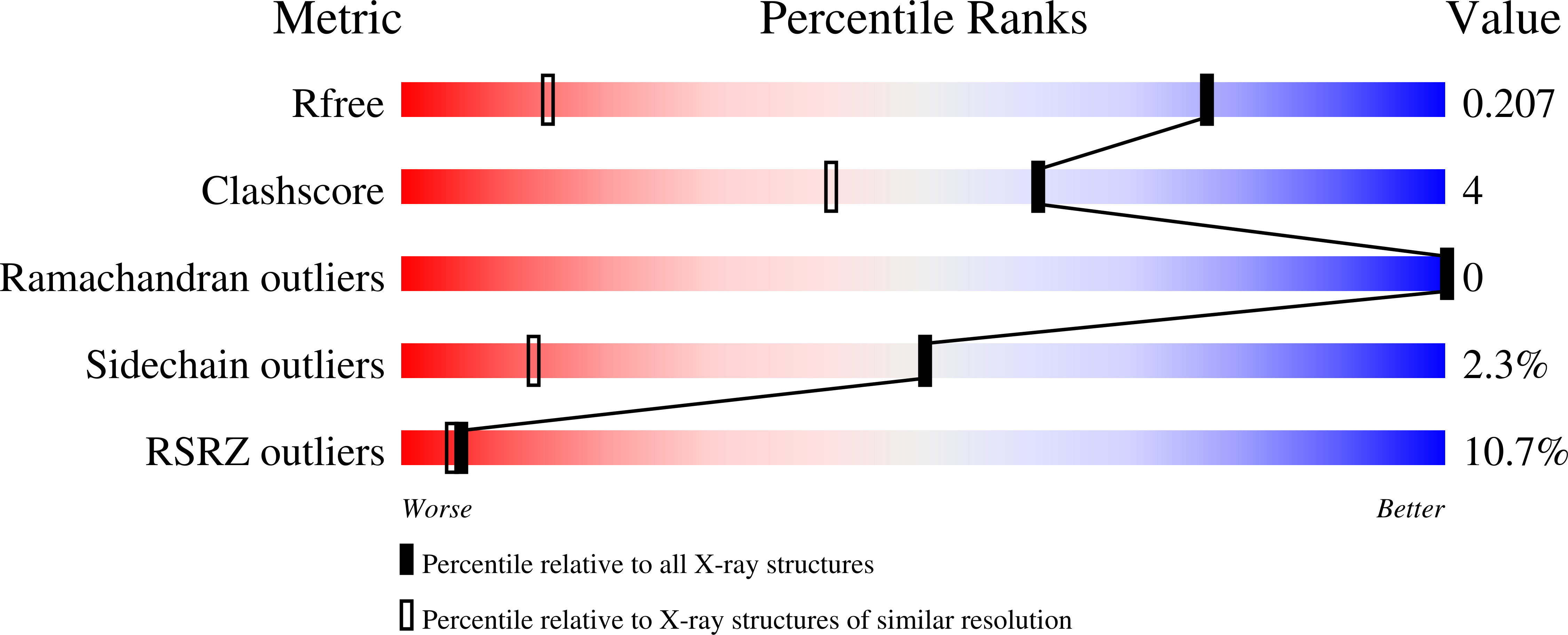
7OVY - PubMed Abstract:
The metalloproteinase ADAM8 is upregulated in several cancers but has a dispensable function under physiological conditions. In tumor cells, ADAM8 is involved in invasion, migration, and angiogenesis. The use of bivalent inhibitors could impair migration and invasion through the double binding to a homodimeric form of ADAM8 located on the cell surface of tumor cells. Herein we report the rational design and synthesis of the first dimeric ADAM8 inhibitors selective over ADAM10 and matrix metalloproteinases. Bivalent derivatives have been obtained by dimerizing the structure of a previously described ADAM17 inhibitor, JG26. In particular, derivative 2 was shown to inhibit ADAM8 proteolytic activity in vitro and in cell-based assays at nanomolar concentration. Moreover, it was more effective than the parent monomeric compound in blocking invasiveness in the breast cancer MDA-MB-231 cell line, thus supporting our hypothesis about the importance of inhibiting the active homodimer of ADAM8.
Organizational Affiliation:
Department of Pharmacy, University of Pisa, via Bonanno 6, 56126 Pisa, Italy.