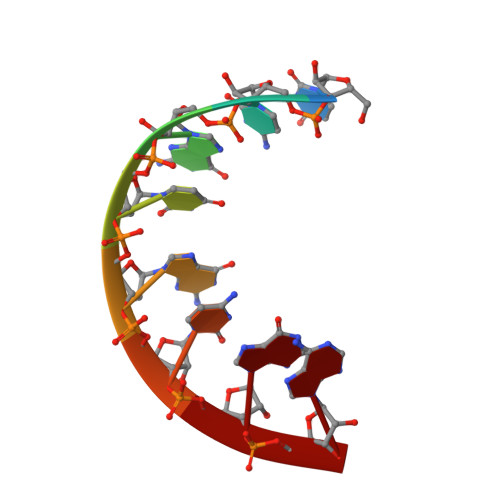
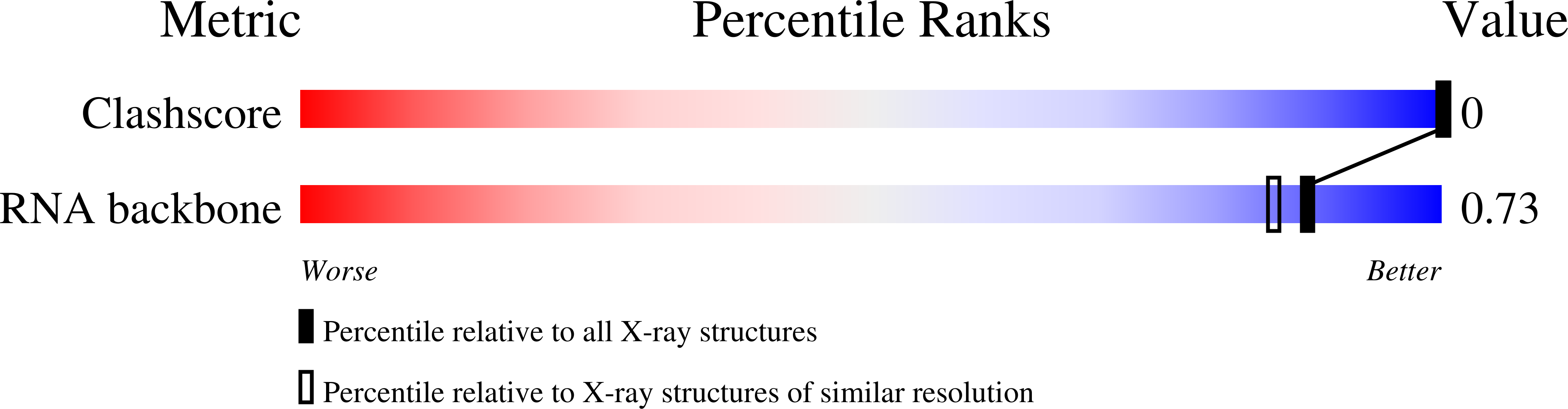Structural Insights Into the 5'UG/3'GU Wobble Tandem in Complex With Ba 2+ Cation.
Ruszkowska, A., Zheng, Y.Y., Mao, S., Ruszkowski, M., Sheng, J.(2021) Front Mol Biosci 8: 762786-762786
- PubMed: 35096964
- DOI: https://doi.org/10.3389/fmolb.2021.762786
- Primary Citation of Related Structures:
7OUO - PubMed Abstract:
G•U wobble base pair frequently occurs in RNA structures. The unique chemical, thermodynamic, and structural properties of the G•U pair are widely exploited in RNA biology. In several RNA molecules, the G•U pair plays key roles in folding, ribozyme catalysis, and interactions with proteins. G•U may occur as a single pair or in tandem motifs with different geometries, electrostatics, and thermodynamics, further extending its biological functions. The metal binding affinity, which is essential for RNA folding, catalysis, and other interactions, differs with respect to the tandem motif type due to the different electrostatic potentials of the major grooves. In this work, we present the crystal structure of an RNA 8-mer duplex r[UCGUGCGA] 2 , providing detailed structural insights into the tandem motif I (5'UG/3'GU) complexed with Ba 2+ cation. We compare the electrostatic potential of the presented motif I major groove with previously published structures of tandem motifs I, II (5'GU/3'UG), and III (5'GG/3'UU). A local patch of a strongly negative electrostatic potential in the major groove of the presented structure forms the metal binding site with the contributions of three oxygen atoms from the tandem. These results give us a better understanding of the G•U tandem motif I as a divalent metal binder, a feature essential for RNA functions.
Organizational Affiliation:
Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland.