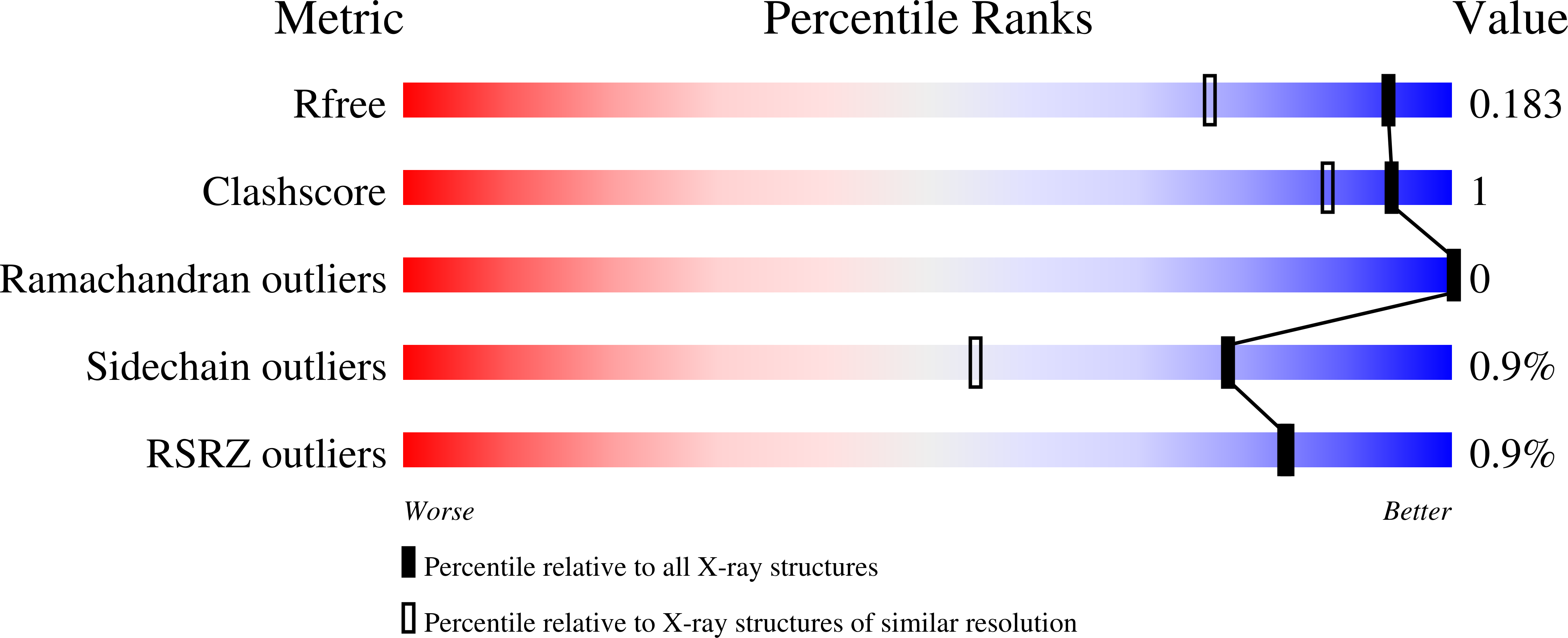
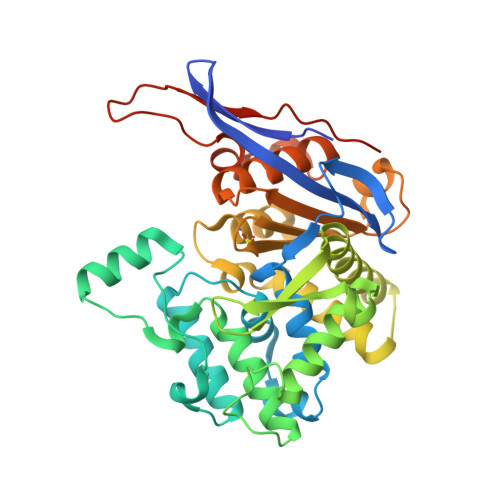
Crystal structures of the elusive Rhizobium etli L-asparaginase reveal a peculiar active site.
Loch, J.I., Imiolczyk, B., Sliwiak, J., Wantuch, A., Bejger, M., Gilski, M., Jaskolski, M.(2021) Nat Commun 12: 6717-6717
- PubMed: 34795296
- DOI: https://doi.org/10.1038/s41467-021-27105-x
- Primary Citation of Related Structures:
7OS3, 7OS5, 7OS6, 7OU1, 7OZ6 - PubMed Abstract:
Rhizobium etli, a nitrogen-fixing bacterial symbiont of legume plants, encodes an essential L-asparaginase (ReAV) with no sequence homology to known enzymes with this activity. High-resolution crystal structures of ReAV show indeed a structurally distinct, dimeric enzyme, with some resemblance to glutaminases and β-lactamases. However, ReAV has no glutaminase or lactamase activity, and at pH 9 its allosteric asparaginase activity is relatively high, with K m for L-Asn at 4.2 mM and k cat of 438 s -1 . The active site of ReAV, deduced from structural comparisons and confirmed by mutagenesis experiments, contains a highly specific Zn 2+ binding site without a catalytic role. The extensive active site includes residues with unusual chemical properties. There are two Ser-Lys tandems, all connected through a network of H-bonds to the Zn center, and three tightly bound water molecules near Ser48, which clearly indicate the catalytic nucleophile.
Organizational Affiliation:
Department of Crystal Chemistry and Crystal Physics, Faculty of Chemistry, Jagiellonian University, Krakow, Poland.