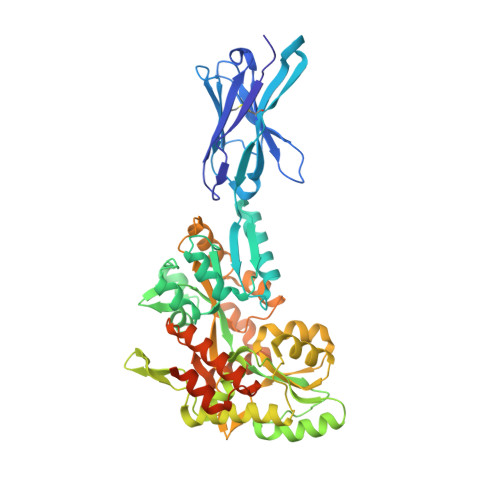
Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Botte, M., Ni, D., Schenck, S., Zimmermann, I., Chami, M., Bocquet, N., Egloff, P., Bucher, D., Trabuco, M., Cheng, R.K.Y., Brunner, J.D., Seeger, M.A., Stahlberg, H., Hennig, M.(2022) Nat Commun 13: 1826-1826
- PubMed: 35383177
- DOI: https://doi.org/10.1038/s41467-022-29459-2
- Primary Citation of Related Structures:
7OMM, 7OMT - PubMed Abstract:
Lipopolysaccharides are major constituents of the extracellular leaflet in the bacterial outer membrane and form an effective physical barrier for environmental threats and for antibiotics in Gram-negative bacteria. The last step of LPS insertion via the Lpt pathway is mediated by the LptD/E protein complex. Detailed insights into the architecture of LptDE transporter complexes have been derived from X-ray crystallography. However, no structure of a laterally open LptD transporter, a transient state that occurs during LPS release, is available to date. Here, we report a cryo-EM structure of a partially opened LptDE transporter in complex with rigid chaperones derived from nanobodies, at 3.4 Å resolution. In addition, a subset of particles allows to model a structure of a laterally fully opened LptDE complex. Our work offers insights into the mechanism of LPS insertion, provides a structural framework for the development of antibiotics targeting LptD and describes a highly rigid chaperone scaffold to enable structural biology of challenging protein targets.
Organizational Affiliation:
leadXpro AG, Park Innovaare, 5234, Villigen, Switzerland.